The rate-limiting step in the folding of a large ribozyme without kinetic traps
- PMID: 12084911
- PMCID: PMC124294
- DOI: 10.1073/pnas.142288399
The rate-limiting step in the folding of a large ribozyme without kinetic traps
Abstract
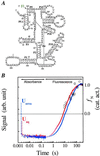
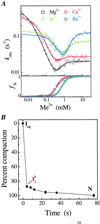
A fundamental question in RNA folding is the nature of the rate-limiting step. Folding of large RNAs often is trapped by the need to undo misfolded structures, which precludes the study of the other, potentially more interesting aspects in the rate-limiting step, such as conformational search, metal ion binding, and the role of productive intermediates. The catalytic domain of the Bacillus subtilis RNase P RNA folds without a kinetic trap, thereby providing an ideal system to elucidate these steps. We analyzed the folding kinetics by using fluorescence and absorbance spectroscopies, catalytic activity, and synchrotron small-angle x-ray scattering. Folding begins with the rapid formation of early intermediates wherein the majority of conformational search occurs, followed by the slower formation of subsequent intermediates. Before the rate-limiting step, more than 98% of the total structure has formed. The rate-limiting step is a small-scale structural rearrangement involving prebound metal ions.
Figures


References
-
- Cole P E, Crothers D M. Biochemistry. 1972;11:4368–4374. - PubMed
-
- Webb P K, Fresco J R. J Mol Biol. 1973;74:387–402. - PubMed
-
- Lynch D C, Schimmel P R. Biochemistry. 1974;13:1841–1852. - PubMed
-
- Zarrinkar P P, Williamson J R. Science. 1994;265:918–924. - PubMed
-
- Banerjee A R, Turner D H. Biochemistry. 1995;34:6504–6512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

