Topological determinants of protein folding
- PMID: 12084924
- PMCID: PMC124342
- DOI: 10.1073/pnas.122076099
Topological determinants of protein folding
Abstract
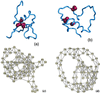
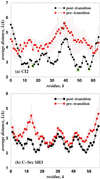
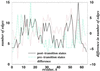
The folding of many small proteins is kinetically a two-state process that represents overcoming the major free-energy barrier. A kinetic characteristic of a conformation, its probability to descend to the native state domain in the amount of time that represents a small fraction of total folding time, has been introduced to determine to which side of the free-energy barrier a conformation belongs. However, which features make a protein conformation on the folding pathway become committed to rapidly descending to the native state has been a mystery. Using two small, well characterized proteins, CI2 and C-Src SH3, we show how topological properties of protein conformations determine their kinetic ability to fold. We use a macroscopic measure of the protein contact network topology, the average graph connectivity, by constructing graphs that are based on the geometry of protein conformations. We find that the average connectivity is higher for conformations with a high folding probability than for those with a high probability to unfold. Other macroscopic measures of protein structural and energetic properties such as radius of gyration, rms distance, solvent-accessible surface area, contact order, and potential energy fail to serve as predictors of the probability of a given conformation to fold.
Figures
References
-
- Fersht A R. Curr Opin Struct Biol. 1995;5:79–84. - PubMed
-
- Mirny L A, Shakhnovich E I. Annu Rev Biophys Biomol Struct. 2001;30:361–396. - PubMed
-
- Du R, Pande V S, Grosberg A Y, Tanaka T, Shakhnovich E I. J Chem Phys. 1998;108:334–350.
-
- Dokholyan N V, Buldyrev S V, Stanley H E, Shakhnovich E I. J Mol Biol. 2000;296:1183–1188. - PubMed
-
- Vendruscolo M, Paci E, Dobson C, Karplus M. Nature (London) 2001;409:641–645. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

