Disease surveillance in recombining pathogens: multilocus genotypes identify sources of human Coccidioides infections
- PMID: 12084944
- PMCID: PMC124424
- DOI: 10.1073/pnas.132178099
Disease surveillance in recombining pathogens: multilocus genotypes identify sources of human Coccidioides infections
Abstract
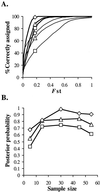
Molecular surveillance of pathogenic microbes works by genotyping isolates with DNA fingerprinting techniques and then using these genotypes to assign individuals to populations. Clonality is assumed in many fingerprinting studies, although this assumption has been shown to be false for many organisms. To accommodate recombining organisms into surveillance programs, methods using population allele frequencies in combination with individual multilocus genotypes are necessary. Here, we develop a statistical method appropriate for haploid recombining microbes that allows individuals to be assigned to populations. We illustrate the usefulness of this technique by inferring the source populations for Coccidioides isolates recovered from patients treated outside the endemic area of Coccidioides sp., the etiological agents of human coccidioidomycosis, but with a travel history including visits to one or more endemic areas.
Figures

References
-
- Daszak P, Cunningham A A, Hyatt D. Science. 2000;287:443–449. - PubMed
-
- Rippon J W. Medical Mycology. Philadelphia: Saunders; 1988.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

