A new way to rapidly create functional, fluorescent fusion proteins: random insertion of GFP with an in vitro transposition reaction
- PMID: 12086589
- PMCID: PMC117241
- DOI: 10.1186/1471-2202-3-7
A new way to rapidly create functional, fluorescent fusion proteins: random insertion of GFP with an in vitro transposition reaction
Abstract
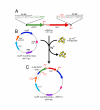
Background: The jellyfish green fluorescent protein (GFP) can be inserted into the middle of another protein to produce a functional, fluorescent fusion protein. Finding permissive sites for insertion, however, can be difficult. Here we describe a transposon-based approach for rapidly creating libraries of GFP fusion proteins.
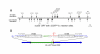
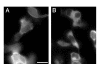
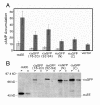
Results: We tested our approach on the glutamate receptor subunit, GluR1, and the G protein subunit, alphas. All of the in-frame GFP insertions produced a fluorescent protein, consistent with the idea that GFP will fold and form a fluorophore when inserted into virtually any domain of another protein. Some of the proteins retained their signaling function, and the random nature of the transposition process revealed permissive sites for insertion that would not have been predicted on the basis of structural or functional models of how that protein works.
Conclusion: This technique should greatly speed the discovery of functional fusion proteins, genetically encodable sensors, and optimized fluorescence resonance energy transfer pairs.
Figures

References
-
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–5. - PubMed
-
- Marshall J, Molloy R, Moss GW, Howe JR, Hughes TE. The jellyfish green fluorescent protein: a new tool for studying ion channel expression and function. Neuron. 1995;14:211–5. - PubMed
-
- Chalfie M. Green fluorescent protein. Photochem Photobiol. 1995;62:651–6. - PubMed
-
- Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

