Overlapping of MINK and CHRNE gene loci in the course of mammalian evolution
- PMID: 12087176
- PMCID: PMC117062
- DOI: 10.1093/nar/gkf407
Overlapping of MINK and CHRNE gene loci in the course of mammalian evolution
Abstract
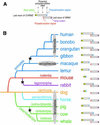
Overlapping of genes, especially in an anti-parallel fashion, is quite rare in eukaryotic genomes. We have found a rare instance of exon overlapping involving CHRNE and MINK gene loci on chromosome 17 in humans. CHRNE codes for the epsilon subunit of the nicotinic acetylcholine receptor (AChRepsilon) whereas MINK encodes a serine/threonine kinase belonging to the GCK family. To elucidate the evolutionary trail of this gene overlapping event, we examined the genomes of a number of primates and found that mutations in the polyadenylation signal of the CHRNE gene in early hominoids led to the overlap. Upon extending this analysis to genomes of other orders of placental mammals, we observed that the overlapping occurred at least three times independently during the course of mammalian evolution. Because CHRNE and MINK are differentially expressed, the potentially hazardous mutations responsible for the exon overlap seem to have escaped evolutionary pressures by differential temporo-spatial expression of the two genes.
Figures


References
-
- Dunham I., Shimizu,N., Roe,B.A., Chissoe,S., Hunt,A.R., Collins,J.E., Bruskiewich,R., Beare,D.M., Clamp,M., Smink,L.J. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. - PubMed
-
- Vanhee-Brossollet C. and Vaquero,C. (1998) Do natural antisense transcripts make sense in eukaryotes? Gene, 211, 1–9. - PubMed
-
- Miyajima N., Horiuchi,R., Shibuya,Y., Fukushige,S., Matsubara,K., Toyoshima,K. and Yamamoto,T. (1989) Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell, 57, 31–39. - PubMed
-
- Batshake B. and Sundelin,J. (1996) The mouse genes for the EP1 prostanoid receptor and the PKN protein kinase overlap. Biochem. Biophys. Res. Commun., 227, 70–76. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

