Analysis of cell-cycle gene expression in Saccharomyces cerevisiae using microarrays and multiple synchronization methods
- PMID: 12087178
- PMCID: PMC117069
- DOI: 10.1093/nar/gkf414
Analysis of cell-cycle gene expression in Saccharomyces cerevisiae using microarrays and multiple synchronization methods
Abstract
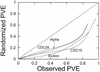
Microarray analysis of gene expression during the yeast division cycle has led to the proposal that a significant number of genes in Saccharomyces cerevisiae are expressed in a cell-cycle-specific manner. Four different methods of synchronization were used for cell-cycle analysis. Randomized data exhibit periodic patterns of lesser strength than the experimental data. Thus the cyclicities in the expression measurements in the four experiments presented do not arise from chance fluctuations or noise in the data. However, when the degree of cyclicity for genes in different experiments are compared, a large degree of non-reproducibility is found. Re-examining the phase timing of peak expression, we find that three of the experiments (those using alpha-factor, CDC28 and CDC15 synchronization) show consistent patterns of phasing, but the elutriation synchrony results demonstrate a different pattern from the other arrest-release synchronization methods. Specific genes can show a wide range of cyclical behavior between different experiments; a gene with high cyclicity in one experiment can show essentially no cyclicity in another experiment. The elutriation experiment, possibly being the least perturbing of the four synchronization methods, may give the most accurate characterization of the state of gene expression during the normal, unperturbed cell cycle. Under this alternative explanation, the observed cyclicities in the other three experiments are a stress response to synchronization, and may not reproduce in unperturbed cells.
Figures



References
-
- Cho R.J., Campbell,M.J., Winzeler,E.A., Steinmetz,L., Conway,A., Wodicka,L., Wolfsberg,T.G., Gabrielian,A.E., Landsman,D., Lockhart,D.J. et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. - PubMed
-
- Aach J., Rindone,W. and Church,G.M. (2000) Systematic management and analysis of yeast gene expression data. Genome Res., 10, 431–445. - PubMed
-
- Aach J. and Church,G.M. (2001) Aligning gene expression time series with time warping algorithms. Bioinformatics, 17, 495–508. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

