On the epigenetic regulation of the human reelin promoter
- PMID: 12087179
- PMCID: PMC117056
- DOI: 10.1093/nar/gkf401
On the epigenetic regulation of the human reelin promoter
Abstract
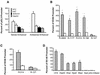
Reln mRNA and protein levels are reduced by approximately 50% in various cortical structures of post-mortem brain from patients diagnosed with schizophrenia or bipolar illness with psychosis. To study mechanisms responsible for this down-regulation, we have analyzed the promoter of the human reelin gene. We show that the reelin promoter directs expression of a reporter construct in multiple human cell types: neuroblastoma cells (SHSY5Y), neuronal precursor cells (NT2), differentiated neurons (hNT) and hepatoma cells (HepG2). Deletion constructs confirmed the presence of multiple elements regulating Reln expression, although the promoter activity is promiscuous, i.e. activity did not correlate with expression of the endogenous gene as reflected in terms of reelin mRNA levels. Co-transfection of the -514 bp human reelin promoter with either Sp1 or Tbr1 demonstrated that these transcription factors activate reporter expression by 6- and 8.5-fold, respectively. Within 400 bp of the RNA start site there are 100 potential CpG targets for DNA methylation. Retinoic acid (RA)-induced differentiation of NT2 cells to hNT neurons was accompanied by increased reelin expression and by the appearance of three DNase I hypersensitive sites 5' to the RNA start site. RA-induced differentiation was also associated with demethylation of the reelin promoter. To test if methylation silenced reelin expression, we methylated the promoter in vitro prior to transfection. In addition, we treated NT2 cells with the methylation inhibitor aza-2'-deoxycytidine and observed a 60-fold increase in reelin mRNA levels. The histone deacetylase inhibitors trichostatin A (TSA) and valproic acid also induced expression of the endogenous reelin promoter, although TSA was considerably more potent. These findings indicate that one determinant responsible for regulating reelin expression is the methylation status of the promoter. Our data also raise the interesting possibility that the down-regulation of reelin expression documented in psychiatric patients might be the consequence of inappropriate promoter hypermethylation.
Figures



References
-
- Impagnatiello F., Guidotti,A.R., Pesold,C., Dwivedi,Y., Caruncho,H., Pisu,M.G., Uzunov,D.P., Smalheiser,N.R., Davis,J.M., Pandey,G.N., Pappas,G.D., Tueting,P., Sharma,R.P. and Costa,E. (1998) A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc. Natl Acad. Sci. USA, 95, 15718–15723. - PMC - PubMed
-
- Costa E., Davis,J., Grayson,D.R., Guidotti,A., Pappas,G.D. and Pesold,C. (2001) Dendritic spine hypoplasia and downregulation of reelin and GABAergic tone in schizophrenia vulnerability. Neurobiol. Dis., 8, 723–742. - PubMed
-
- Guidotti A., Auta,J., Davis,J., Dwivedi,Y., Grayson,D.R., Impagnatiello,F., Pandey,G., Pesold,C., Sharma,R., Uzunov,D. and Costa,E. (2000) Reelin and GAD67 expression is decreased in postmortem brain of schizophrenia and bipolar disorder patients. Arch. Gen. Psychiatr., 57, 1061–1069. - PubMed
-
- Selemon L.D. and Goldman-Rakic,P.S. (1999) The reduced neuropil hypothesis: a circuit based model of schizophrenia. Biol. Psychiatr., 45, 17–25. - PubMed
-
- Fatemi S.H., Earle,J.A. and McMenomy,T. (2000) Reduction in reelin immunoreactivity in hippocampus of subjects with schizophrenia, bipolar disorder and major depression. Mol. Psychiatr., 5, 654–663. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

