Parameter optimized surfaces (POPS): analysis of key interactions and conformational changes in the ribosome
- PMID: 12087181
- PMCID: PMC117037
- DOI: 10.1093/nar/gkf373
Parameter optimized surfaces (POPS): analysis of key interactions and conformational changes in the ribosome
Abstract
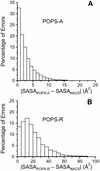
We present a new method for the calculation of solvent accessible surface areas at the atomic and residue levels, which we call parameter optimized surfaces (POPS-A and POPS-R ). Atomic and residue areas (the latter simulated with a single sphere centered at the C(alpha)s atom for amino acids and at the P atom for nucleotides) have been optimized versus accurate all-atoms methods. We concentrated on an analytical formula for the approximation of solvent accessibilities. The formula is simple, easily derivable and fast to compute, therefore it is practical for use in molecular dynamics simulations as an approximation to the first solvation shell. The residue based approach POPS-R has been derived as a useful tool for the analysis of large macromolecular assemblies like the ribosome, and is especially suited for use in refinement of low resolution structures. The structures of the 70S, 50S and 30S ribosomes have been analyzed in detail and most of the interactions within the subunits and at their interfaces were clearly identified. Some interesting differences between 30S alone and within the 70S have been highlighted. Owing to the presence of the P-tRNA in the 70S ribosome, localized conformational rearrangements occur within the subunits, exposing Arg and Lys residues to negatively charged binding sites of P-tRNA. POPS-R also allows for estimates of the loss of free energy of solvation upon complex formation, particularly useful in designing new protein-RNA complexes and in suggesting more focused experimental work.
Figures




References
-
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
-
- Doudna J. (2000) Structural genomics of DNA. Nature Struct. Biol., Suppl 7, 954–956. - PubMed
-
- Marcotte E., Pellegrini,M., Thompson,M., Yeates,T. and Eisenberg,D. (1999) A combined algorithm for genome-wide prediction of protein function. Nature, 402, 83–86. - PubMed
-
- Thornton J., Todd,A., Milburn,D., Borkakoti,N. and Orengo,C. (2000) From structure to function: approaches and limitations. Nature Struct. Biol., Suppl 7, 991–994. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

