A pH-jump approach for investigating secondary structure refolding kinetics in RNA
- PMID: 12087188
- PMCID: PMC117070
- DOI: 10.1093/nar/gnf057
A pH-jump approach for investigating secondary structure refolding kinetics in RNA
Abstract
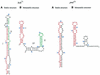
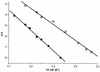
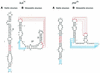
It has been shown that premature translation of the plasmid-mediated toxin in hok/sok of plasmid R1 and pnd/pndB of plasmid R483 is prevented during transcription of the hok and pnd mRNAs by the formation of metastable hairpins at the 5'-end of the mRNA. Here, an experimental approach is presented, which allows the accurate measurement of the refolding kinetics of the 5'-end RNA fragments in vitro without chemically modifying the RNA. The method is based on acid denaturation followed by a pH-jump to neutral pH as a novel way to trap kinetically favoured RNA secondary structures, allowing the measurement of a wide range of biologically relevant refolding rates, with or without the use of standard stopped-flow equipment. The refolding rates from the metastable to the stable conformation in both the hok74 and pnd58 5'-end RNA fragments were determined by using UV absorbance changes corresponding to the structural rearrangements. The measured energy barriers showed that the refolding path does not need complete unfolding of the metastable structures before the formation of the final structures. Two alternative models of such a pathway are discussed.
Figures



Similar articles
-
Metastable structures and refolding kinetics in hok mRNA of plasmid R1.RNA. 1999 Nov;5(11):1408-18. doi: 10.1017/s1355838299990805. RNA. 1999. PMID: 10580469 Free PMC article.
-
Programmed cell death by hok/sok of plasmid R1: coupled nucleotide covariations reveal a phylogenetically conserved folding pathway in the hok family of mRNAs.J Mol Biol. 1997 Oct 17;273(1):26-37. doi: 10.1006/jmbi.1997.1295. J Mol Biol. 1997. PMID: 9367743
-
The hok mRNA family.RNA Biol. 2012 Dec;9(12):1399-404. doi: 10.4161/rna.22746. Epub 2012 Dec 1. RNA Biol. 2012. PMID: 23324554
-
The hok killer gene family in gram-negative bacteria.New Biol. 1990 Nov;2(11):946-56. New Biol. 1990. PMID: 2101633 Review.
-
Antisense RNA-regulated programmed cell death.Annu Rev Genet. 1997;31:1-31. doi: 10.1146/annurev.genet.31.1.1. Annu Rev Genet. 1997. PMID: 9442888 Review.
Cited by
-
Kinetic mechanism of conformational switch between bistable RNA hairpins.J Am Chem Soc. 2012 Aug 1;134(30):12499-507. doi: 10.1021/ja3013819. Epub 2012 Jul 19. J Am Chem Soc. 2012. PMID: 22765263 Free PMC article.
-
Kinetic Mechanism of RNA Helix-Terminal Basepairing-A Kinetic Minima Network Analysis.Biophys J. 2019 Nov 5;117(9):1674-1683. doi: 10.1016/j.bpj.2019.09.017. Epub 2019 Sep 20. Biophys J. 2019. PMID: 31590890 Free PMC article.
-
Structural parameters affecting the kinetics of RNA hairpin formation.Nucleic Acids Res. 2006 Jul 19;34(12):3568-76. doi: 10.1093/nar/gkl445. Print 2006. Nucleic Acids Res. 2006. PMID: 16855293 Free PMC article.
-
Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism.RNA. 2007 Dec;13(12):2202-12. doi: 10.1261/rna.635307. Epub 2007 Oct 24. RNA. 2007. PMID: 17959930 Free PMC article.
-
Understanding the kinetic mechanism of RNA single base pair formation.Proc Natl Acad Sci U S A. 2016 Jan 5;113(1):116-21. doi: 10.1073/pnas.1517511113. Epub 2015 Dec 22. Proc Natl Acad Sci U S A. 2016. PMID: 26699466 Free PMC article.
References
-
- Gerdes K., Gultyaev,A.P., Franch,T., Pedersen,K. and Mikkelsen,N.D. (1997) Antisense RNA-regulated programmed cell death. Annu. Rev. Genet., 31, 1–31. - PubMed
-
- Lodmell J.S. and Dahlberg,A.E. (1997) A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science, 277, 1262–1267. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

