Modulation of HIV-1 replication by RNA interference
- PMID: 12087358
- PMCID: PMC9524216
- DOI: 10.1038/nature00896
Modulation of HIV-1 replication by RNA interference
Abstract
RNA interference (RNAi) is the process by which double-stranded RNA (dsRNA) directs sequence-specific degradation of messenger RNA in animal and plant cells. In mammalian cells, RNAi can be triggered by 21-nucleotide duplexes of small interfering RNA (siRNA). Here we describe inhibition of early and late steps of HIV-1 replication in human cell lines and primary lymphocytes by siRNAs targeted to various regions of the HIV-1 genome. We demonstrate that synthetic siRNA duplexes or plasmid-derived siRNAs inhibit HIV-1 infection by specifically degrading genomic HIV-1 RNA, thereby preventing formation of viral complementary-DNA intermediates. These results demonstrate the utility of RNAi for modulating the HIV replication cycle and provide evidence that genomic HIV-1 RNA, as it exists within a nucleoprotein reverse-transcription complex, is amenable to siRNA-mediated degradation.
Conflict of interest statement
Competing interests statement
The authors declare that they have no competing financial interests.
Figures

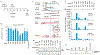
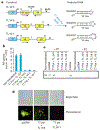
Comment in
-
Medicine: silencing viruses with RNA.Nature. 2002 Jul 25;418(6896):379-80. doi: 10.1038/418379a. Nature. 2002. PMID: 12140542 No abstract available.
References
-
- Sharp PA RNA interference–2001. Genes Dev. 15, 485–490 (2001). - PubMed
-
- Hutvagner G & Zamore PD RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev 12, 225–232 (2002). - PubMed
-
- Elbashir SM et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001). - PubMed
-
- Fire A et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). - PubMed
-
- Zamore PD, Tuschl T, Sharp PA & Bartel DP RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

