Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families
- PMID: 12087515
- PMCID: PMC379167
- DOI: 10.1086/341944
Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families
Abstract
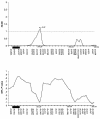
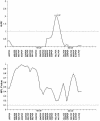
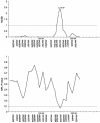
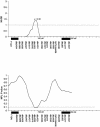
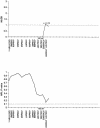
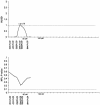
Cleft lip with or without cleft palate (CL/P) is a common congenital anomaly. Birth prevalences range from 1/500 to 1/1,000 and are consistently higher in Asian populations than in populations of European descent. Therefore, it is of interest to determine whether the CL/P etiological factors in Asian populations differ from those in white populations. A sample of 36 multiplex families were ascertained through probands with CL/P who were from Shanghai. This is the first reported genome-scan study of CL/P in any Asian population. Genotyping of Weber Screening Set 9 (387 short tandem-repeat polymorphisms with average spacing approximately 9 cM [range 1-19 cM]) was performed by the Mammalian Genotyping Service of Marshfield Laboratory. Presented here are the results for the 366 autosomal markers. Linkage between each marker and CL/P was assessed by two-point and multipoint LOD scores, as well as with multipoint heterogeneity LOD scores (HLODs) plus model-free identity-by-descent statistics and the multipoint NPL statistic. In addition, association was assessed via the transmission/disequilibrium test. LOD-score and HLOD calculations were performed under a range of models of inheritance of CL/P. The following regions had positive multipoint results (HLOD > or =1.0 and/or NPL P< or =.05): chromosomes 1 (90-110 cM), 2 (220-250 cM), 3 (130-150 cM), 4 (140-170 cM), 6 (70-100 cM), 18 (110 cM), and 21 (30-50 cM). The most significant multipoint linkage results (HLOD > or =2.0; alpha=0.37) were for chromosomes 3q and 4q. Associations with P< or =.05 were found for loci on chromosomes 3, 5-7, 9, 11, 12, 16, 20, and 21. The most significant association result (P=.009) was found with D16S769 (51 cM).
Figures


Similar articles
-
Genome-wide and interaction linkage scan for nonsyndromic cleft lip with or without cleft palate in two multiplex families in Shenyang, China.Biomed Environ Sci. 2010 Oct;23(5):363-70. doi: 10.1016/S0895-3988(10)60077-3. Biomed Environ Sci. 2010. PMID: 21112484
-
Genome-scan for loci involved in cleft lip with or without cleft palate in consanguineous families from Turkey.Am J Med Genet A. 2004 Apr 15;126A(2):111-22. doi: 10.1002/ajmg.a.20564. Am J Med Genet A. 2004. PMID: 15057975
-
Genome scan for loci involved in nonsyndromic cleft lip with or without cleft palate in families from West Bengal, India.Am J Med Genet A. 2004 Oct 15;130A(3):265-71. doi: 10.1002/ajmg.a.30252. Am J Med Genet A. 2004. PMID: 15378549
-
Chromosome 17: gene mapping studies of cleft lip with or without cleft palate in Chinese families.Cleft Palate Craniofac J. 2003 Jan;40(1):71-9. doi: 10.1597/1545-1569_2003_040_0071_cgmsoc_2.0.co_2. Cleft Palate Craniofac J. 2003. PMID: 12498608
-
Nonsyndromic cleft lip with or without cleft palate in China: assessment of candidate regions.Cleft Palate Craniofac J. 2002 Mar;39(2):149-56. doi: 10.1597/1545-1569_2002_039_0149_nclwow_2.0.co_2. Cleft Palate Craniofac J. 2002. PMID: 11879070
Cited by
-
Analysis of large phenotypic variability of EEC and SHFM4 syndromes caused by K193E mutation of the TP63 gene.PLoS One. 2012;7(5):e35337. doi: 10.1371/journal.pone.0035337. Epub 2012 May 4. PLoS One. 2012. PMID: 22574117 Free PMC article.
-
Genetic determinants of facial clefting: analysis of 357 candidate genes using two national cleft studies from Scandinavia.PLoS One. 2009;4(4):e5385. doi: 10.1371/journal.pone.0005385. Epub 2009 Apr 29. PLoS One. 2009. PMID: 19401770 Free PMC article.
-
Autosomal dominant nonsyndromic cleft lip and palate: significant evidence of linkage at 18q21.1.Am J Hum Genet. 2007 Jul;81(1):180-8. doi: 10.1086/518944. Epub 2007 May 18. Am J Hum Genet. 2007. PMID: 17564975 Free PMC article.
-
Feasibility of identifying families for genetic studies of birth defects using the National Health Interview Survey.BMC Public Health. 2004 May 12;4:16. doi: 10.1186/1471-2458-4-16. BMC Public Health. 2004. PMID: 15140264 Free PMC article.
-
Discovery of candidate genes for nonsyndromic cleft lip palate through genome-wide linkage analysis of large extended families in the Malay population.BMC Genet. 2016 Feb 11;17:39. doi: 10.1186/s12863-016-0345-x. BMC Genet. 2016. PMID: 26868259 Free PMC article.
References
Electronic-Database Information
-
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for Weber Screening Set 9)
-
- Electronic Scholarly Publishing, http://www.esp.org/books/darwin/variation/facsimile/ (for facsimile of revised second edition of Darwin's The Variation of Animals and Plants under Domestication)
-
- Genetics Research Group, School of Dental Medicine, University of Pittsburgh, http://www.sdmgenetics.pitt.edu (for markers and allele sizes and frequencies)
References
-
- Bateson W (1909) Mendel's principles of hereditary. Cambridge University Press, Cambridge
-
- Bear JC (1976) A genetic study of facial clefting in northern England. Clin Genet 9:277–284 - PubMed
-
- Carinci F, Pezzetti F, Scapoli L, Martinelli M, Carinci P, Tognon M (2000) Genetics of nonsyndromic cleft lip and palate: a review of international studies and data regarding the Italian population. Cleft Palate Craniofac J 37:33–40 - PubMed
-
- Carter CO (1976) Genetics of common single malformations. Br Med Bull 32:21–26 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

