Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces
- PMID: 12089021
- PMCID: PMC126800
- DOI: 10.1128/AEM.68.7.3401-3407.2002
Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces
Abstract
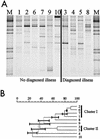
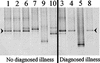
The human gastrointestinal (GI) tract harbors a complex community of bacterial cells in the mucosa, lumen, and feces. Since most attention has been focused on bacteria present in feces, knowledge about the mucosa-associated bacterial communities in different parts of the colon is limited. In this study, the bacterial communities in feces and biopsy samples from the ascending, transverse, and descending colons of 10 individuals were analyzed by using a 16S rRNA approach. Flow cytometric analysis indicated that 10(5) to 10(6) bacteria were present in the biopsy samples. To visualize the diversity of the predominant and the Lactobacillus group community, denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA gene amplicons was performed. DGGE analysis and similarity index comparisons demonstrated that the predominant mucosa-associated bacterial community was host specific and uniformly distributed along the colon but significantly different from the fecal community (P < 0.01). The Lactobacillus group-specific profiles were less complex than the profiles reflecting the predominant community. For 6 of the 10 individuals the community of Lactobacillus-like bacteria in the biopsy samples was similar to that in the feces. Amplicons having 99% sequence similarity to the 16S ribosomal DNA of Lactobacillus gasseri were detected in the biopsy samples of nine individuals. No significant differences were observed between healthy and diseased individuals. The observed host-specific DGGE profiles of the mucosa-associated bacterial community in the colon support the hypothesis that host-related factors are involved in the determination of the GI tract microbial community.
Figures


Similar articles
-
Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon.Appl Environ Microbiol. 2003 Dec;69(12):7545-8. doi: 10.1128/AEM.69.12.7545-7548.2003. Appl Environ Microbiol. 2003. PMID: 14660412 Free PMC article.
-
Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples.Appl Environ Microbiol. 2007 Nov;73(22):7435-42. doi: 10.1128/AEM.01143-07. Epub 2007 Sep 21. Appl Environ Microbiol. 2007. PMID: 17890331 Free PMC article.
-
Molecular characterization of the bacteria adherent to human colorectal mucosa.J Appl Microbiol. 2006 Mar;100(3):460-9. doi: 10.1111/j.1365-2672.2005.02783.x. J Appl Microbiol. 2006. PMID: 16478485
-
DGGE and 16S rDNA sequencing analysis of bacterial communities in colon content and feces of pigs fed whole crop rice.Anaerobe. 2007 Jun-Aug;13(3-4):127-33. doi: 10.1016/j.anaerobe.2007.03.001. Epub 2007 Mar 12. Anaerobe. 2007. PMID: 17446093
-
Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals.Gastroenterology. 2019 Oct;157(4):1081-1092.e3. doi: 10.1053/j.gastro.2019.05.068. Epub 2019 Jun 5. Gastroenterology. 2019. PMID: 31175864
Cited by
-
Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals.Sci Rep. 2020 Sep 11;10(1):14977. doi: 10.1038/s41598-020-71939-2. Sci Rep. 2020. PMID: 32917913 Free PMC article. Clinical Trial.
-
Dietary inhibitors of histone deacetylases in intestinal immunity and homeostasis.Front Immunol. 2013 Aug 1;4:226. doi: 10.3389/fimmu.2013.00226. eCollection 2013. Front Immunol. 2013. PMID: 23914191 Free PMC article.
-
Role of commensal gut bacteria in inflammatory bowel diseases.Gut Microbes. 2012 Nov-Dec;3(6):544-55. doi: 10.4161/gmic.22156. Epub 2012 Oct 11. Gut Microbes. 2012. PMID: 23060017 Free PMC article. Review.
-
Stool sampling and DNA isolation kits affect DNA quality and bacterial composition following 16S rRNA gene sequencing using MiSeq Illumina platform.Sci Rep. 2019 Sep 25;9(1):13837. doi: 10.1038/s41598-019-49520-3. Sci Rep. 2019. PMID: 31554833 Free PMC article.
-
Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract.Appl Environ Microbiol. 2004 May;70(5):2791-800. doi: 10.1128/AEM.70.5.2791-2800.2004. Appl Environ Microbiol. 2004. PMID: 15128534 Free PMC article.
References
-
- Alander, M., R. Korpela, M. Saxelin, T. Vilpponen-Salmela, T. Mattila-Sandholm, and A. von Wright. 1997. Recovery of Lactobacillus rhamnosus GG from human colonic biopsies. Lett. Appl. Microbiol. 24:361-364. - PubMed
-
- Bry, L., P. G. Falk, T. Midtvedt, and J. I. Gordon. 1996. A model of host-microbial interactions in an open mammalian ecosystem. Science 273:1381-1383. - PubMed
-
- Gibson, G. R., J. H. Cummings, and G. T. Macfarlane. 1991. Growth and activities of sulphate-reducing bacteria in the gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol. Ecol. 86:103-112.
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

