Characterization of an unusual Mycobacterium: a possible missing link between Mycobacterium marinum and Mycobacterium ulcerans
- PMID: 12089250
- PMCID: PMC120612
- DOI: 10.1128/JCM.40.7.2370-2380.2002
Characterization of an unusual Mycobacterium: a possible missing link between Mycobacterium marinum and Mycobacterium ulcerans
Abstract
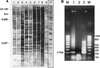
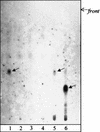
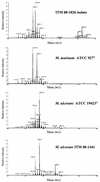
In an attempt to characterize an unusual mycobacterial isolate from a 44-year-old patient living in France, we applied phenotypic characterizations and various previously described molecular methods for the taxonomic classification of mycobacteria. The results of the investigations were compared to those obtained in a previous study with a set of temporally and geographically diverse Mycobacterium ulcerans (n = 29) and Mycobacterium marinum (n = 29) isolates (K. Chemlal, G. Huys, P.-A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels, J. Clin. Microbiol. 39:3272-3278, 2001). The isolate, designated ITM 00-1026 (IPP 2000-372), is closely related to M. marinum according to its phenotypic properties, lipid pattern, and partial 16S rRNA sequence. Moreover, fingerprinting by amplified fragment length polymorphism (AFLP) analysis unequivocally classified this strain as a member of the species M. marinum, although it lacked two species-specific AFLP marker bands. However, PCR and restriction fragment length polymorphism analysis based on M. ulcerans-specific insertion sequence IS2404 showed the presence of this element in a low copy number in isolate ITM 00-1026. In conclusion, the designation of this isolate as a transitional species further supports the recent claim by Stinear et al. (T. Stinear, G. Jenkin, P. D. Johnson, and J. K. Davies, J. Bacteriol. 182:6322-6330, 2000) that M. ulcerans represents a relatively recent phylogenetic derivative of M. marinum resulting from the systematic acquisition of foreign DNA fragments.
Figures





References
-
- Aronson, J. D. 1926. Spontaneous tuberculosis in salt water fish. Infect. Dis. 39:315-320.
-
- Bhatt, A., and T. Kieser. 1999. Transposition of IS117 of Streptomyces coelicolor A3(2) in Mycobacterium smegmatis. Microbiology 145:1201-1207. - PubMed
-
- Chemlal, K., K. De Ridder, P. A. Fonteyne. W. M. Meyers, J. Swings, and F. Portaels. 2001. The use of IS2404 restriction fragment length polymorphism suggests the diversity of Mycobacterium ulcerans from different geographical areas. Am. J. Trop. Med. Hyg. 64:270-273. - PubMed
-
- Chemlal, K., G. Huys, P.-A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels. 2001. Evaluation of PCR restriction profile analysis, IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 39:3272-3278. - PMC - PubMed
-
- Daffé, M., M.-A. Lanéelle, and C. Lacave. 1991. Structure and stereochemistry of mycolic acids of Mycobacterium marinum and Mycobacterium ulcerans. Res. Microbiol. 142:397-403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

