Highly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA
- PMID: 12089255
- PMCID: PMC120571
- DOI: 10.1128/JCM.40.7.2408-2419.2002
Highly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA
Abstract
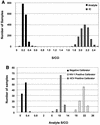
Various nucleic acid assays have been developed and implemented for diagnostics and therapeutic monitoring of human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) infections. The high-throughput, semiautomated assays described here were developed to provide a method suitable for screening plasma specimens for the presence of HIV-1 and HCV RNAs. Three assays were developed: a multiplex HIV-1/HCV assay for simultaneous detection of HIV-1 and HCV, and discriminatory assays for specific detection of HIV-1 and HCV. The assay systems utilize three proprietary technologies: (i) target capture-based sample preparation, (ii) transcription-mediated amplification (TMA), and (iii) hybridization protection assay (HPA). An internal control is incorporated into each reaction to control for every step of the assay and identify random false-negative reactions. The assays demonstrated a sensitivity of at least 100 copies/ml for each target, and they detected with similar sensitivity all major variants of HCV and HIV-1, including HIV-1 group O strains. Assay sensitivity for one virus was not affected by the presence of the other. The specificity of these TMA-driven assays was >or=99.5% in both normal donor specimens and plasma containing potentially interfering substances or other blood-borne pathogens. Statistical receiver operating characteristic plots of 1 - specificity versus sensitivity data determined very wide analyte cutoff values for each assay at the point at which the assay specificity and sensitivity were both >or=99.5%. The sensitivity, specificity, and throughput capability predict that these assays will be valuable for large-volume plasma screening, either in a blood bank setting or in other diagnostic applications.
Figures


Similar articles
-
Evaluation of a new molecular assay for detection of human immunodeficiency virus type 1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA.J Clin Virol. 2006 Jul;36(3):166-76. doi: 10.1016/j.jcv.2005.12.003. Epub 2006 Jan 20. J Clin Virol. 2006. PMID: 16427802
-
Sensitivity of the Procleix HIV-1/HCV assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA in a high-risk population.J Clin Microbiol. 2002 Jul;40(7):2387-91. doi: 10.1128/JCM.40.7.2387-2391.2002. J Clin Microbiol. 2002. PMID: 12089252 Free PMC article.
-
Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA.J Clin Microbiol. 2001 Aug;39(8):2937-45. doi: 10.1128/JCM.39.8.2937-2945.2001. J Clin Microbiol. 2001. PMID: 11474017 Free PMC article.
-
Mini review: current molecular methods for the detection and quantification of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1.Int J Infect Dis. 2014 Aug;25:145-9. doi: 10.1016/j.ijid.2014.04.007. Epub 2014 Jun 10. Int J Infect Dis. 2014. PMID: 24927665 Review.
-
[Detection of the nucleic acids of hepatitis B and C viruses and human immunodeficiency virus for the biological screening of blood donations. Viral Hepatitis and Retrovirus Working Groups and Subgroup for Molecular Biology Applied to Transfusion Virology of the French Blood Transfusion Society].Transfus Clin Biol. 1998 Apr;5(2):139-46. doi: 10.1016/s1246-7820(98)80004-9. Transfus Clin Biol. 1998. PMID: 9618839 Review. French.
Cited by
-
The Babesia observational antibody (BAOBAB) study: A cross-sectional evaluation of Babesia in two communities in Kilosa district, Tanzania.PLoS Negl Trop Dis. 2019 Aug 14;13(8):e0007632. doi: 10.1371/journal.pntd.0007632. eCollection 2019 Aug. PLoS Negl Trop Dis. 2019. PMID: 31412024 Free PMC article.
-
Sensitive detection assays for influenza RNA do not reveal viremia in US blood donors.J Infect Dis. 2012 Mar 15;205(6):886-94. doi: 10.1093/infdis/jir863. Epub 2012 Jan 31. J Infect Dis. 2012. PMID: 22293429 Free PMC article.
-
Tools and Techniques for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)/COVID-19 Detection.Clin Microbiol Rev. 2021 May 12;34(3):e00228-20. doi: 10.1128/CMR.00228-20. Print 2021 Jun 16. Clin Microbiol Rev. 2021. PMID: 33980687 Free PMC article. Review.
-
Ensuring the biologic safety of plasma-derived therapeutic proteins: detection, inactivation, and removal of pathogens.BioDrugs. 2005;19(2):79-96. doi: 10.2165/00063030-200519020-00002. BioDrugs. 2005. PMID: 15807628 Free PMC article. Review.
-
Reducing infection transmission in solid organ transplantation through donor nucleic acid testing: a cost-effectiveness analysis.Am J Transplant. 2013 Oct;13(10):2611-8. doi: 10.1111/ajt.12429. Epub 2013 Aug 22. Am J Transplant. 2013. PMID: 24034208 Free PMC article.
References
-
- Abravaya, K., C. Esping, R. Hoenle, J. Gorzowski, R. Perry, P. Kroeger, J. Robinson, and R. Flanders. 2000. Performance of a multiplex qualitative PCR LCx assay for detection of human immunodeficiency virus type 1 (HIV-1) group M subtypes, group O, and HIV-2. J. Clin. Microbiol. 38:716-723. - PMC - PubMed
-
- Allain, J. P. 2000. Genomic screening for blood-borne viruses in transfusion settings. Clin. Lab. Haematol. 22:1-10. - PubMed
-
- Allain, J. P. 2000. Will genome detection replace serology in blood screening for microbial agents? Baillieres Best Pract. Res. Clin. Haematol. 13:615-629. - PubMed
-
- Alter, H. J., R. H. Purcell, J. W. Shih, J. C. Melpolder, M. Houghton, Q. L. Choo, and G. Kuo. 1989. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N. Engl. J. Med. 321:1494-1500. - PubMed
-
- Arnold, L., P. W. Hammond, W. A. Wiese, and N. C. Nelson. 1989. Assay formats involving acridinium-ester-labeled DNA probes. Clin. Chem. 35:1588-1594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

