Minimizing the workup of blood culture contaminants: implementation and evaluation of a laboratory-based algorithm
- PMID: 12089259
- PMCID: PMC120579
- DOI: 10.1128/JCM.40.7.2437-2444.2002
Minimizing the workup of blood culture contaminants: implementation and evaluation of a laboratory-based algorithm
Abstract
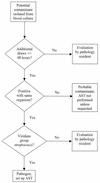
An algorithm was implemented in the clinical microbiology laboratory to assess the clinical significance of organisms that are often considered contaminants (coagulase-negative staphylococci, aerobic and anaerobic diphtheroids, Micrococcus spp., Bacillus spp., and viridans group streptococci) when isolated from blood cultures. From 25 August 1999 through 30 April 2000, 12,374 blood cultures were submitted to the University of Iowa Clinical Microbiology Laboratory. Potential contaminants were recovered from 495 of 1,040 positive blood cultures. If one or more additional blood cultures were obtained within +/-48 h and all were negative, the isolate was considered a contaminant. Antimicrobial susceptibility testing (AST) of these probable contaminants was not performed unless requested. If no additional blood cultures were submitted or there were additional positive blood cultures (within +/-48 h), a pathology resident gathered patient clinical information and made a judgment regarding the isolate's significance. To evaluate the accuracy of these algorithm-based assignments, a nurse epidemiologist in approximately 60% of the cases performed a retrospective chart review. Agreement between the findings of the retrospective chart review and the automatic classification of the isolates with additional negative blood cultures as probable contaminants occurred among 85.8% of 225 isolates. In response to physician requests, AST had been performed on 15 of the 32 isolates with additional negative cultures considered significant by retrospective chart review. Agreement of pathology resident assignment with the retrospective chart review occurred among 74.6% of 71 isolates. The laboratory-based algorithm provided an acceptably accurate means for assessing the clinical significance of potential contaminants recovered from blood cultures.
Figures
References
-
- Archer, G. L. 1985. Coagulase-negative staphylococci in blood cultures: the clinician's dilemma. Infect. Control 6:477-478. - PubMed
-
- Bates, D. W., L. Goldman, and T. H. Lee. 1991. Contaminant blood cultures and resource utilization: the true consequences of false-positive results. JAMA 265:365-369. - PubMed
-
- Bates, D. W., and T. H. Lee. 1992. Rapid classification of positive blood cultures: prospective validation of a multivariate algorithm. JAMA 267:1962-1966. - PubMed
-
- Correa, L., and D. Pittet. 2000. Problems and solutions in hospital-acquired bacteraemia. J. Hosp. Infect. 46:89-95. - PubMed
-
- Dominguez-de Villota, E., A. Algora-Weber, I. Millan, et al. 1987. Early evaluation of coagulase negative staphylococcus in blood samples of intensive care unit patients: a clinically uncertain judgement. Intensive Care Med. 13:390-394. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous