Seminested PCR for diagnosis of candidemia: comparison with culture, antigen detection, and biochemical methods for species identification
- PMID: 12089267
- PMCID: PMC120535
- DOI: 10.1128/JCM.40.7.2483-2489.2002
Seminested PCR for diagnosis of candidemia: comparison with culture, antigen detection, and biochemical methods for species identification
Abstract
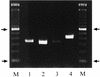
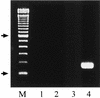
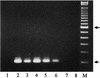
The rapid detection and identification of Candida species in clinical laboratories are extremely important for the management of patients with hematogenous candidiasis. The presently available culture and biochemical methods for detection and species identification of Candida are time-consuming and lack the required sensitivity and specificity. In this study, we have established a seminested PCR (snPCR) using universal and species-specific primers for detection of Candida species in serum specimens. The universal outer primers amplified the 3' end of 5.8S ribosomal DNA (rDNA) and the 5' end of 28S rDNA, including the internally transcribed spacer 2 (ITS2), generating 350- to 410-bp fragments from the four commonly encountered Candida species, viz., C. albicans, C. tropicalis, C. glabrata, and C. parapsilosis. The species-specific primers, complementary to unique sequences within the ITS2 of each test species, amplified species-specific DNA in the reamplification step of the snPCR. The sensitivity of Candida detection by snPCR in spiked serum specimens was close to 1 organism/ml. Evaluation of snPCR for specific identification of Candida species with 76 clinical Candida isolates showed 99% concordant results with the Vitek and/or ID32C yeast identification system. Further evaluation of snPCR for detection of Candida species in sera from culture-proven (n = 12), suspected (n = 16), and superficially colonized (n = 10) patients and healthy subjects (n = 12) showed that snPCR results were consistently negative with sera from healthy individuals and colonized patients. In culture-proven candidemia patients, the snPCR results were in full agreement with blood culture results with respect to both positivity and species identity. In addition, snPCR detected candidemia due to two Candida species in five patients, compared to three by blood culture. In the category of suspected candidemia with negative blood cultures for Candida, nine patients (56%) were positive by snPCR; two of them had dual infection with C. albicans and either C. tropicalis or C. glabrata. In conclusion, the snPCR developed in this study is specific and more sensitive than culture for the detection of Candida species in serum specimens. Moreover, the improved detection of cases of candidemia caused by more than one Candida species is an additional advantage.
Figures

References
-
- Abi-Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24:1122-1128. - PubMed
-
- Botelho, A. R., and R. J. Planta. 1994. Specific identification of Candida albicans by hybridization with oligonucleotides derived from ribosomal DNA internal spacers. Yeast 10:709-717. - PubMed
-
- Cheung, L. L., and J. B. Hudson. 1988. Development of DNA probes for Candida albicans. Diagn. Microbiol. Infect. Dis. 10:171-179. - PubMed
-
- Chryssanthou, E., B. Andersson, B. Petrini, S. Lofdahl, and J. Tollemar. 1994. Detection of Candida albicans DNA in serum by polymerase chain reaction. Scand. J. Infect. Dis. 26:479-485. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

