Detection of Epstein-Barr virus genomes in peripheral blood B cells from solid-organ transplant recipients by fluorescence in situ hybridization
- PMID: 12089275
- PMCID: PMC120580
- DOI: 10.1128/JCM.40.7.2533-2544.2002
Detection of Epstein-Barr virus genomes in peripheral blood B cells from solid-organ transplant recipients by fluorescence in situ hybridization
Abstract
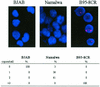
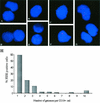
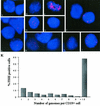
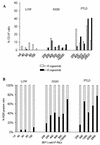
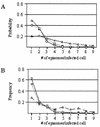
Resolution of Epstein-Barr Virus (EBV) infection in pediatric solid-organ transplant recipients often leads to an asymptomatic carrier state characterized by a persistently elevated circulating EBV load that is 2 to 4 orders of magnitude greater than the load typical of healthy latently infected individuals. Elevated EBV loads in immunosuppressed individuals are associated with an increased risk for development of posttransplant lymphoproliferative disease. We have performed fluorescence in situ hybridization (FISH) studies with peripheral blood B cells from carriers of persistent EBV loads in order to directly quantitate the number of EBV genomes per infected cell. Patients were assigned to two groups on the basis of the level of the persistent load (low-load carriers, 8 to 200 genomes/10(5) peripheral blood lymphocytes; high-load carriers, >200 genomes/10(5) peripheral blood lymphocytes). FISH analysis revealed that the low-load carriers predominantly had circulating virus-infected cells harboring one or two genome copies/cell. High-load carriers also had cells harboring one or two genome copies/cell; in addition, however, they carried a distinct population of cells with high numbers of viral genome copies. The increased viral loads correlated with an increase in the frequency of cells containing high numbers of viral genomes. We conclude that low-load carriers possess EBV-infected cells that are in a state similar to normal latency, whereas high-load carriers possess two populations of virus-positive B cells, one of which carries an increased number of viral genomes per cell and is not typical of normal latency.
Figures

Similar articles
-
Surface immunoglobulin-deficient Epstein-Barr virus-infected B cells in the peripheral blood of pediatric solid-organ transplant recipients.J Clin Microbiol. 2004 Dec;42(12):5802-10. doi: 10.1128/JCM.42.12.5802-5810.2004. J Clin Microbiol. 2004. PMID: 15583315 Free PMC article.
-
Pediatric solid-organ transplant recipients carry chronic loads of Epstein-Barr virus exclusively in the immunoglobulin D-negative B-cell compartment.J Clin Microbiol. 2001 Apr;39(4):1407-15. doi: 10.1128/JCM.39.4.1407-1415.2001. J Clin Microbiol. 2001. PMID: 11283064 Free PMC article.
-
Comparison of six different specimen types for Epstein-Barr viral load quantification in peripheral blood of pediatric patients after heart transplantation or after allogeneic hematopoietic stem cell transplantation.J Clin Virol. 2012 Mar;53(3):186-94. doi: 10.1016/j.jcv.2011.11.010. Epub 2011 Dec 17. J Clin Virol. 2012. PMID: 22182950
-
Epstein-Barr virus load monitoring: its role in the prevention and management of post-transplant lymphoproliferative disease.Transpl Infect Dis. 2001 Jun;3(2):79-87. doi: 10.1034/j.1399-3062.2001.003002079.x. Transpl Infect Dis. 2001. PMID: 11395973 Review.
-
B lymphocytes and Epstein-Barr virus: the lesson of post-transplant lymphoproliferative disorders.Autoimmun Rev. 2007 Dec;7(2):96-101. doi: 10.1016/j.autrev.2007.02.012. Epub 2007 Mar 26. Autoimmun Rev. 2007. PMID: 18035317 Review.
Cited by
-
Aspirin Inhibits Natural Killer/T-Cell Lymphoma by Modulation of VEGF Expression and Mitochondrial Function.Front Oncol. 2019 Jan 14;8:679. doi: 10.3389/fonc.2018.00679. eCollection 2018. Front Oncol. 2019. PMID: 30693272 Free PMC article.
-
Detection of EBV genomes in plasmablasts/plasma cells and non-B cells in the blood of most patients with EBV lymphoproliferative disorders by using Immuno-FISH.Blood. 2010 Nov 25;116(22):4546-59. doi: 10.1182/blood-2010-05-285452. Epub 2010 Aug 10. Blood. 2010. PMID: 20699441 Free PMC article.
-
Surface immunoglobulin-deficient Epstein-Barr virus-infected B cells in the peripheral blood of pediatric solid-organ transplant recipients.J Clin Microbiol. 2004 Dec;42(12):5802-10. doi: 10.1128/JCM.42.12.5802-5810.2004. J Clin Microbiol. 2004. PMID: 15583315 Free PMC article.
-
Evodiamine suppresses endometriosis development induced by early EBV exposure through inhibition of ERβ.Front Pharmacol. 2024 Aug 1;15:1426660. doi: 10.3389/fphar.2024.1426660. eCollection 2024. Front Pharmacol. 2024. PMID: 39148548 Free PMC article.
-
Infection and persistence of rhesus monkey rhadinovirus in immortalized B-cell lines.J Virol. 2006 Apr;80(7):3644-9. doi: 10.1128/JVI.80.7.3644-3649.2006. J Virol. 2006. PMID: 16537632 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

