Physical mapping of genes in somatic cell radiation hybrids by comparative genomic hybridization to cDNA microarrays
- PMID: 12093373
- PMCID: PMC116723
- DOI: 10.1186/gb-2002-3-6-research0026
Physical mapping of genes in somatic cell radiation hybrids by comparative genomic hybridization to cDNA microarrays
Abstract
Background: Somatic cell mutants can be informative in the analysis of a wide variety of cellular processes. The use of map-based positional cloning strategies in somatic cell hybrids to analyze genes responsible for recessive mutant phenotypes is often tedious, however, and remains a major obstacle in somatic cell genetics. To fulfill the need for more efficient gene mapping in somatic cell mutants, we have developed a new DNA microarray comparative genomic hybridization (array-CGH) method that can rapidly and efficiently map the physical location of genes complementing somatic cell mutants to a small candidate genomic region. Here we report experiments that establish the validity and efficacy of the methodology.
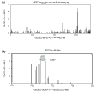
Results: CHO cells deficient for hypoxanthine:guanine phosphoribosyl transferase (HPRT) were fused with irradiated normal human fibroblasts and subjected to HAT selection. Cy5-labeled genomic DNA from the surviving hybrids containing the HPRT gene was mixed with Cy3-labeled genomic DNA from normal CHO cells and hybridized to a microarray containing 40,185 cDNAs, representing 29,399 genes (UniGene clusters). The DNA spots with the highest Cy5:Cy3 fluorescence ratios corresponded to a group of genes mapping within a 1 Mb interval centered near position 142.7 Mb on the X chromosome, the genomic location of HPRT.
Conclusion: The results indicate that our physical mapping method based on radiation hybrids and array-CGH should significantly enhance the speed and efficiency of positional cloning in somatic cell genetics.
Figures



References
-
- Stark GR, Gudkov AV. Forward genetics in mammalian cells: functional approaches to gene discovery. Hum Mol Genet. 1999;8:1925–1938. - PubMed
-
- List of genes cloned by positional cloning http://genome.nhgri.nih.gov/clone
-
- Lin JY, Bedford JS. Regional gene mapping using mixed radiation hybrids and reverse chromosome painting. Radiat Res. 1997;148:405–412. - PubMed
-
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. - PubMed
-
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

