NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp
- PMID: 12093725
- PMCID: PMC126088
- DOI: 10.1093/emboj/cdf339
NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp
Abstract
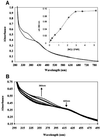
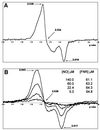
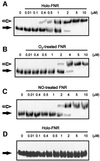
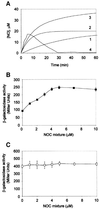
Nitric oxide (NO) is a signalling and defence molecule of major importance in biology. The flavohaemoglobin Hmp of Escherichia coli is involved in protective responses to NO. Because hmp gene transcription is repressed by the O(2)-responsive regulator FNR, we investigated whether FNR also senses NO. The [4Fe-4S](2+) cluster of FNR is oxygen labile and controls protein dimerization and site-specific DNA binding. NO reacts anaerobically with the Fe-S cluster of purified FNR, generating spectral changes consistent with formation of a dinitrosyl-iron-cysteine complex. NO-inactivated FNR can be reconstituted, suggesting physiological relevance. FNR binds at an FNR box within the hmp promoter (P(hmp)). FNR samples inactivated by either O(2) or NO bind specifically to P(hmp), but with lower affinity. Dose-dependent up-regulation of P(hmp) in vivo by NO concentrations of pathophysiological relevance is abolished by fnr mutation, and NO also modulates expression from model FNR-regulated promoters. Thus, FNR can respond to not only O(2), but also NO, with major implications for global gene regulation in bacteria. We propose an NO-mediated mechanism of hmp regulation by which E.coli responds to NO challenge.
Figures




References
-
- Bouton C., Hirling,H. and Drapier,J.-C. (1997) Modulation of iron regulatory protein functions. Further insights into the role of nitrogen- and oxygen-derived reactive species. J. Biol. Chem., 272, 19969–19975. - PubMed
-
- Chung M.H., Kasai,H., Jones,D.S., Inoue,H., Ishikawa,H., Otsuka,E. and Nishimura,S. (1991) An endonuclease activity of Escherichia coli that specifically removes 8-hydroxyguanine residues from DNA. Mutat. Res., 254, 1–12. - PubMed
-
- Costanzo S., Ménage,S., Purrello,R., Bonomo,R.P. and Fontecave M. (2001) Re-examination of the formation of dinitrosyl-iron complexes during reaction of S-nitrosothiols with Fe(II). Inorg. Chim. Acta, 318, 1–7.
-
- Crawford W.J. and Goldberg,D.E. (1998) Regulation of the Salmonella typhimurium flavohemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J. Biol. Chem., 273, 34028–34032. - PubMed
-
- Cruz-Ramos H., Hughes,M.N. and Poole,R.K. (2000) Nitric oxide is an effector in bacterial gene expression mediated by FNR and FNR-like proteins. In Moncada,S., Gustafsson,L.E., Wiklund,N.P. and Higgs,E.A (eds), The Biology of Nitric Oxide Part 7. Portland Press, London, UK, p. 158.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

