Structural analysis of two enzymes catalysing reverse metabolic reactions implies common ancestry
- PMID: 12093726
- PMCID: PMC126076
- DOI: 10.1093/emboj/cdf298
Structural analysis of two enzymes catalysing reverse metabolic reactions implies common ancestry
Abstract
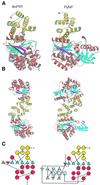
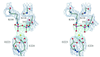
The crystal structure of the dimeric anthranilate phosphoribosyltransferase (AnPRT) reveals a new category of phosphoribosyltransferases, designated as class III. The active site of this enzyme is located within the flexible hinge region of its two-domain structure. The pyrophosphate moiety of phosphoribosylpyrophosphate is co-ordinated by a metal ion and is bound by two conserved loop regions within this hinge region. With the structure of AnPRT available, structural analysis of all enzymatic activities of the tryptophan biosynthesis pathway is complete, thereby connecting the evolution of its enzyme members to the general development of metabolic processes. Its structure reveals it to have the same fold, topology, active site location and type of association as class II nucleoside phosphorylases. At the level of sequences, this relationship is mirrored by 13 structurally invariant residues common to both enzyme families. Taken together, these data imply common ancestry of enzymes catalysing reverse biological processes--the ribosylation and deribosylation of metabolic pathway intermediates. These relationships establish new links for enzymes involved in nucleotide and amino acid metabolism.
Figures





References
-
- Bauerle R., Hess,J. and French,S. (1987) Anthranilate synthase– anthranilate phosphoribosyltransferase complex and subunits of Salmonella typhimurium. Methods Enzymol., 142, 366–386. - PubMed
-
- Brünger A.T. (1992) The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–474. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Chaudhuri B.N., Lange,S.C., Myers,R.S., Chittur,S.V., Davisson,V.J. and Smith,J.L. (2001) Crystal structure of imidazole glycerol phosphate synthase. A tunnel through a (β/α)(8) barrel joins two active sites. Structure, 9, 987–997. - PubMed
-
- Cohen G.E. (1997) A program to superimpose protein coordinates, accounting for insertions and deletions. J. Appl. Crystallogr., 30, 1160–1161.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

