Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection
- PMID: 12093732
- PMCID: PMC125391
- DOI: 10.1093/emboj/cdf325
Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection
Abstract
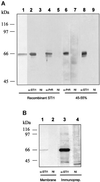
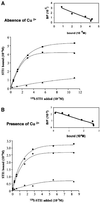
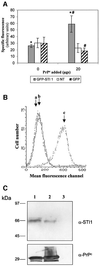
Prions are composed of an isoform of a normal sialoglycoprotein called PrP(c), whose physiological role has been under investigation, with focus on the screening for ligands. Our group described a membrane 66 kDa PrP(c)-binding protein with the aid of antibodies against a peptide deduced by complementary hydropathy. Using these antibodies in western blots from two-dimensional protein gels followed by sequencing the specific spot, we have now identified the molecule as stress-inducible protein 1 (STI1). We show that this protein is also found at the cell membrane besides the cytoplasm. Both proteins interact in a specific and high affinity manner with a K(d) of 10(-7) M. The interaction sites were mapped to amino acids 113-128 from PrP(c) and 230-245 from STI1. Cell surface binding and pull-down experiments showed that recombinant PrP(c) binds to cellular STI1, and co-immunoprecipitation assays strongly suggest that both proteins are associated in vivo. Moreover, PrP(c) interaction with either STI1 or with the peptide we found that represents the binding domain in STI1 induce neuroprotective signals that rescue cells from apoptosis.
Figures




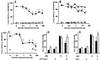
References
-
- Aguzzi A. and Weissmann,C. (1997) Prion research: the next frontiers. Nature, 389, 795–798. - PubMed
-
- Ausubel F.M., Brent,K., Struhl,K., Kingston,R.E., Moore,D.D., Seidman,J.G. and Smith,J.A. (eds) (1993) Current Protocols in Molecular Biology. Wiley Interscience, New York.
-
- Beck K., Hunter,I. and Engel,J. (1990) Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J., 4, 148–160. - PubMed
-
- Blatch G.L., Lassle,M., Takatori,T., Grandhi,T., Kundra,V. and Zetter,B.R. (1995) Molecular characterization of extendin: a protein localized in extending pseudopodia. Proc. Am. Assoc. Cancer Res., 36, 68.
-
- Blatch G.L., Lassle,M., Zetter,B.R. and Kundra,V. (1997) Isolation of a mouse cDNA encoding mSTI1, a stress-inducible protein containing the TPR motif. Gene, 194, 277–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

