An embryonic/fetal beta-type globin gene repressor contains a nuclear receptor TR2/TR4 heterodimer
- PMID: 12093744
- PMCID: PMC126089
- DOI: 10.1093/emboj/cdf340
An embryonic/fetal beta-type globin gene repressor contains a nuclear receptor TR2/TR4 heterodimer
Abstract
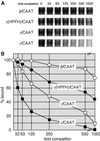
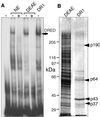
We recently described an erythroid epsilon-globin gene repressor activity, which we named DRED (direct repeat erythroid-definitive). We show that DRED binds with high affinity to DR1 sites in the human embryonic (epsilon-) and fetal (gamma-) globin gene promoters, but the adult beta-globin promoter has no DR1 element. DRED is a 540 kDa complex; sequence determination showed that it contains the nuclear orphan receptors TR2 and TR4. TR2 and TR4 form a heterodimer that binds to the epsilon and gamma promoter DR1 sites. One mutation in a DR1 site causes elevated gamma-globin transcription in human HPFH (hereditary persistence of fetal hemoglobin) syndrome, and we show that this mutation reduces TR2/TR4 binding in vitro. The two receptor mRNAs are expressed at all stages of murine and human erythropoiesis; their forced transgenic expression reduces endogenous embryonic epsilony-globin transcription. These data suggest that TR2/TR4 forms the core of a larger DRED complex that represses embryonic and fetal globin transcription in definitive erythroid cells, and therefore that inhibition of its activity might be an attractive intervention point for treating sickle cell anemia.
Figures





References
-
- Ayer D.E., Lawrence,Q.A. and Eisenman,R.N. (1995) Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell, 80, 767–776. - PubMed
-
- Barberis A., Superti-Furga,G. and Busslinger,M. (1987) Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone promoter. Cell, 50, 347–359. - PubMed
-
- Behe M.J. and Englander,S.W. (1979) Mixed gelation theory. Kinetics, equilibrium and gel incorporation in sickle hemoglobin mixtures. J. Mol. Biol., 133, 137–160. - PubMed
-
- Chang C. and Kokontis,J. (1988) Identification of a new member of the steroid receptor super-family by cloning and sequence analysis. Biochem. Biophys. Res. Commun., 155, 971–977. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

