The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability
- PMID: 12093753
- PMCID: PMC126085
- DOI: 10.1093/emboj/cdf335
The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability
Abstract
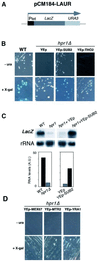
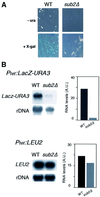
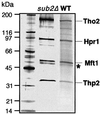
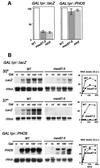
The THO complex is a multimeric factor containing four polypeptides, Tho2, Hpr1, Mft1 and Thp2. Mutations in any of the genes encoding THO confer impairment of transcription and a transcription-dependent hyper-recombination phenotype, suggesting that THO has a functional role in gene expression. Using an in vivo assay developed to study expression of long and G+C-rich DNA sequences, we have isolated SUB2, a gene involved in mRNA splicing and export, as a multicopy suppressor of the gene expression defect of hpr1 Delta. Further investigation of a putative functional relationship between mRNA metabolism and THO revealed that mRNA export mutants sub2, yra1, mex67 and mtr2 have similar defective transcription and hyper-recombination phenotypes as THO mutants. In addition, THO becomes essential in cells with a defective Mex67 mRNA export er. Finally, we have shown that THO has the ability to associate with RNA and DNA in vitro. These results indicate a functional link between the processes of elongation and metabolism of nascent mRNA mediated by THO and mRNA export proteins, which have important consequences for the maintenance of genome stability.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

