Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process
- PMID: 12093756
- PMCID: PMC126079
- DOI: 10.1093/emboj/cdf326
Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process
Abstract
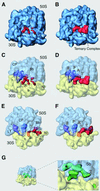
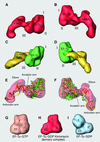
During the elongation cycle of protein biosynthesis, the specific amino acid coded for by the mRNA is delivered by a complex that is comprised of the cognate aminoacyl-tRNA, elongation factor Tu and GTP. As this ternary complex binds to the ribosome, the anticodon end of the tRNA reaches the decoding center in the 30S subunit. Here we present the cryo- electron microscopy (EM) study of an Escherichia coli 70S ribosome-bound ternary complex stalled with an antibiotic, kirromycin. In the cryo-EM map the anticodon arm of the tRNA presents a new conformation that appears to facilitate the initial codon-anticodon interaction. Furthermore, the elbow region of the tRNA is seen to contact the GTPase-associated center on the 50S subunit of the ribosome, suggesting an active role of the tRNA in the transmission of the signal prompting the GTP hydrolysis upon codon recognition.
Figures
Similar articles
-
Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy.Nat Struct Biol. 2003 Nov;10(11):899-906. doi: 10.1038/nsb1003. Epub 2003 Oct 19. Nat Struct Biol. 2003. PMID: 14566331
-
Ensemble cryo-EM elucidates the mechanism of translation fidelity.Nature. 2017 Jun 1;546(7656):113-117. doi: 10.1038/nature22397. Epub 2017 May 24. Nature. 2017. PMID: 28538735 Free PMC article.
-
Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.Nature. 2020 Aug;584(7822):640-645. doi: 10.1038/s41586-020-2447-x. Epub 2020 Jul 1. Nature. 2020. PMID: 32612237 Free PMC article.
-
Structural dynamics of ribosomal RNA during decoding on the ribosome.Biochimie. 2002 Aug;84(8):745-54. doi: 10.1016/s0300-9084(02)01409-8. Biochimie. 2002. PMID: 12457562 Review.
-
Alignment/misalignment hypothesis for tRNA selection by the ribosome.Biochimie. 2006 Aug;88(8):1075-89. doi: 10.1016/j.biochi.2006.07.002. Epub 2006 Jul 26. Biochimie. 2006. PMID: 16890341 Review.
Cited by
-
Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM.Nature. 2015 Apr 23;520(7548):567-70. doi: 10.1038/nature14275. Epub 2015 Feb 23. Nature. 2015. PMID: 25707802
-
A twisted tRNA intermediate sets the threshold for decoding.RNA. 2003 Apr;9(4):384-5. doi: 10.1261/rna.2184703. RNA. 2003. PMID: 12649490 Free PMC article.
-
Ribosomal protein L3: gatekeeper to the A site.Mol Cell. 2007 Mar 23;25(6):877-88. doi: 10.1016/j.molcel.2007.02.015. Mol Cell. 2007. PMID: 17386264 Free PMC article.
-
Irreversible chemical steps control intersubunit dynamics during translation.Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15364-9. doi: 10.1073/pnas.0805299105. Epub 2008 Sep 29. Proc Natl Acad Sci U S A. 2008. PMID: 18824686 Free PMC article.
-
Non-Conserved Residues in Clostridium acetobutylicum tRNA(Ala) Contribute to tRNA Tuning for Efficient Antitermination of the alaS T Box Riboswitch.Life (Basel). 2015 Sep 28;5(4):1567-82. doi: 10.3390/life5041567. Life (Basel). 2015. PMID: 26426057 Free PMC article.
References
-
- Abdulkarim F., Liljas,L. and Hughes,D. (1994) Mutations to kirromycin resistance occur in the interface of domains I and II of EF-Tu–GTP. FEBS Lett., 352, 118–122. - PubMed
-
- Abel K.M., Yoder,M.D., Hilgenfeld,R. and Jurnak,F. (1996) An α to β conformational switch in EF-Tu. Structure, 4, 1153–1159. - PubMed
-
- Agrawal R.K., Heagle,A.B., Penzeck,P., Grassucci,R.A. and Frank,J. (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol., 6, 643–647. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

