The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium
- PMID: 12093901
- PMCID: PMC123171
- DOI: 10.1073/pnas.132181499
The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium
Abstract
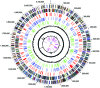
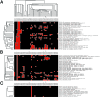
The complete genome of the green-sulfur eubacterium Chlorobium tepidum TLS was determined to be a single circular chromosome of 2,154,946 bp. This represents the first genome sequence from the phylum Chlorobia, whose members perform anoxygenic photosynthesis by the reductive tricarboxylic acid cycle. Genome comparisons have identified genes in C. tepidum that are highly conserved among photosynthetic species. Many of these have no assigned function and may play novel roles in photosynthesis or photobiology. Phylogenomic analysis reveals likely duplications of genes involved in biosynthetic pathways for photosynthesis and the metabolism of sulfur and nitrogen as well as strong similarities between metabolic processes in C. tepidum and many Archaeal species.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

