The fifth essential DNA polymerase phi in Saccharomyces cerevisiae is localized to the nucleolus and plays an important role in synthesis of rRNA
- PMID: 12093911
- PMCID: PMC123106
- DOI: 10.1073/pnas.142277999
The fifth essential DNA polymerase phi in Saccharomyces cerevisiae is localized to the nucleolus and plays an important role in synthesis of rRNA
Abstract
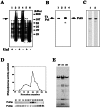
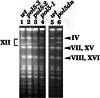
We report that POL5 encodes the fifth essential DNA polymerase in Saccharomyces cerevisiae. Pol5p was identified and purified from yeast cell extracts and is an aphidicolin-sensitive DNA polymerase that is stimulated by yeast proliferating cell nuclear antigen (PCNA). Thus, we named Pol5p DNA polymerase phi. Temperature-sensitive pol5-1-- -3 mutants did not arrest at G(2)/M at the restrictive temperature. Furthermore, the polymerase active-site mutant POL5dn gene complements the lethality of Delta pol5. These results suggest that the polymerase activity of Pol5p is not required for the in vivo function of Pol5p. rRNA synthesis was severely inhibited at the restrictive temperature in the temperature-sensitive pol5-3 mutant cells, suggesting that an essential function of Pol5p is rRNA synthesis. Pol5p is localized exclusively to the nucleolus and binds near or at the enhancer region of rRNA-encoding DNA repeating units.
Figures
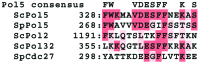


Similar articles
-
Pol5p, a novel binding partner to Cdc10p in fission yeast involved in rRNA production.Mol Genet Genomics. 2006 Oct;276(4):391-401. doi: 10.1007/s00438-006-0144-6. Epub 2006 Jul 1. Mol Genet Genomics. 2006. PMID: 16816948
-
Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system.EMBO J. 2002 Oct 15;21(20):5498-507. doi: 10.1093/emboj/cdf539. EMBO J. 2002. PMID: 12374750 Free PMC article.
-
A polymerase switch in the synthesis of rRNA in Saccharomyces cerevisiae.Mol Cell Biol. 1995 May;15(5):2420-8. doi: 10.1128/MCB.15.5.2420. Mol Cell Biol. 1995. PMID: 7739526 Free PMC article.
-
DNA polymerase II, the epsilon polymerase of Saccharomyces cerevisiae.Prog Nucleic Acid Res Mol Biol. 1993;46:93-120. doi: 10.1016/s0079-6603(08)61019-3. Prog Nucleic Acid Res Mol Biol. 1993. PMID: 8234788 Review. No abstract available.
-
DNA protein interactions at the rRNA of Saccharomyces cerevisiae.Ital J Biochem. 2007 Jun;56(2):81-90. Ital J Biochem. 2007. PMID: 17722648 Review.
Cited by
-
Vibrio vulnificus induces the death of a major bacterial species in the mouse gut via cyclo-Phe-Pro.Microbiome. 2021 Jul 20;9(1):161. doi: 10.1186/s40168-021-01095-w. Microbiome. 2021. PMID: 34284824 Free PMC article.
-
A non-radioactive DNA synthesis assay demonstrates that elements of the Sigma 1278b Mip1 mitochondrial DNA polymerase domain and C-terminal extension facilitate robust enzyme activity.Yeast. 2021 Apr;38(4):262-275. doi: 10.1002/yea.3541. Epub 2021 Jan 26. Yeast. 2021. PMID: 33270277 Free PMC article.
-
Epigeneitc silencing of ribosomal RNA genes by Mybbp1a.J Biomed Sci. 2012 Jun 11;19(1):57. doi: 10.1186/1423-0127-19-57. J Biomed Sci. 2012. PMID: 22686419 Free PMC article.
-
Pol5 is an essential ribosome biogenesis factor required for 60S ribosomal subunit maturation in Saccharomyces cerevisiae.RNA. 2019 Nov;25(11):1561-1575. doi: 10.1261/rna.072116.119. Epub 2019 Aug 14. RNA. 2019. PMID: 31413149 Free PMC article.
-
Assembly and nuclear export of pre-ribosomal particles in budding yeast.Chromosoma. 2014 Aug;123(4):327-44. doi: 10.1007/s00412-014-0463-z. Epub 2014 May 11. Chromosoma. 2014. PMID: 24817020 Review.
References
-
- Kawasaki Y, Sugino A. Mol Cells. 2001;12:277–285. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

