Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1
- PMID: 12093920
- PMCID: PMC123153
- DOI: 10.1073/pnas.152147499
Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1
Abstract
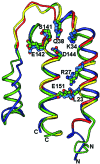
Human IL-10 (hIL-10) modulates critical immune and inflammatory responses by way of interactions with its high- (IL-10R1) and low-affinity (IL-10R2) cell surface receptors. Human cytomegalovirus exploits the IL-10 signaling pathway by expressing a functional viral IL-10 homolog (cmvIL-10), which shares only 27% sequence identity with hIL-10 yet signals through IL-10R1 and IL-10R2. To define the molecular basis of this virus-host interaction, we determined the 2.7-A crystal structure of cmvIL-10 bound to the extracellular fragment of IL-10R1 (sIL-10R1). The structure reveals cmvIL-10 forms a disulfide-linked homodimer that binds two sIL-10R1 molecules. Although cmvIL-10 and hIL-10 share similar intertwined topologies and sIL-10R1 binding sites, their respective interdomain angles differ by approximately 40 degrees. This difference results in a striking re-organization of the IL-10R1s in the putative cell surface complex. Solution binding studies show cmvIL-10 and hIL-10 share essentially identical affinities for sIL-10R1 whereas the Epstein-Barr virus IL-10 homolog (ebvIL-10), whose structure is highly similar to hIL-10, exhibits a approximately 20-fold reduction in sIL-10R1 affinity. Our results suggest cmvIL-10 and ebvIL-10 have evolved different molecular mechanisms to engage the IL-10 receptors that ultimately enhance the respective ability of their virus to escape immune detection.
Figures



References
-
- Moore K W, de Waal Malefyt R, Coffman R L, O'Garra A. Annu Rev Immunol. 2001;19:683–765. - PubMed
-
- Liu Y, Wei S H-Y, Ho A S-Y, de Waal Malefyt R, Moore K W. J Immunol. 1994;152:1821–1929. - PubMed
-
- Walter M R, Nagabhushan T L. Biochemistry. 1995;34:12118–12125. - PubMed
-
- Zdanov A, Schalk-Hihi C, Gustchina A, Tsang M, Weatherbee J, Wlodawer A. Structure (London) 1995;3:591–601. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

