Polyvalent cation receptor proteins (CaRs) are salinity sensors in fish
- PMID: 12093923
- PMCID: PMC123123
- DOI: 10.1073/pnas.152294399
Polyvalent cation receptor proteins (CaRs) are salinity sensors in fish
Abstract
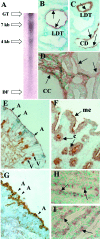
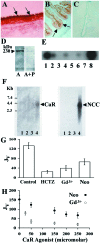
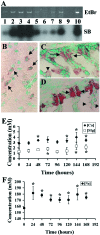
To determine whether calcium polyvalent cation-sensing receptors (CaRs) are salinity sensors in fish, we used a homology-based cloning strategy to isolate a 4.1-kb cDNA encoding a 1,027-aa dogfish shark (Squalus acanthias) kidney CaR. Expression studies in human embryonic kidney cells reveal that shark kidney senses combinations of Ca(2+), Mg(2+), and Na(+) ions at concentrations present in seawater and kidney tubules. Shark kidney is expressed in multiple shark osmoregulatory organs, including specific tubules of the kidney, rectal gland, stomach, intestine, olfactory lamellae, gill, and brain. Reverse transcriptase-PCR amplification using specific primers in two teleost fish, winter flounder (Pleuronectes americanus) and Atlantic salmon (Salmo salar), reveals a similar pattern of CaR tissue expression. Exposure of the lumen of winter flounder urinary bladder to the CaR agonists, Gd(3+) and neomycin, reversibly inhibit volume transport, which is important for euryhaline teleost survival in seawater. Within 24-72 hr after transfer of freshwater-adapted Atlantic salmon to seawater, there are increases in their plasma Ca(2+), Mg(2+), and Na(+) that likely serve as a signal for internal CaRs, i.e., brain, to sense alterations in salinity in the surrounding water. We conclude that CaRs act as salinity sensors in both teleost and elasmobranch fish. Their tissue expression patterns in fish provide insights into CaR functions in terrestrial animals including humans.
Figures


References
-
- Brown E M, Pollack M D, Seidman C E, Seidman J G, Chou Y-H W, Riccardi D, Hebert S C. N Engl J Med. 1995;333:234–240. - PubMed
-
- Rogers K V, Dunn C K, Hebert S C, Brown E M. Brain Res. 1997;744:47–56. - PubMed
-
- Chattopadhyay N, Cheng I, Rogers K, Riccardi D, Hall A, Diaz R, Hebert S C, Soybel D I, Brown E M. Am J Physiol. 1998;274:G122–G130. - PubMed
-
- Quinn S J, Kifor O, Trivedi S, Diaz R, Vassilev P, Brown E. J Biol Chem. 1998;273:19579–19586. - PubMed
-
- Baum M A, Harris H W. Am J Med Sci. 1998;316:321–328. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

