Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model
- PMID: 12097338
- PMCID: PMC186630
- DOI: 10.1101/gr.228902
Computation-based discovery of related transcriptional regulatory modules and motifs using an experimentally validated combinatorial model
Abstract
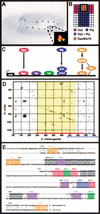
Gene expression is regulated by transcription factors that interact with cis-regulatory elements. Predicting these elements from sequence data has proven difficult. We describe here a successful computational search for elements that direct expression in a particular temporal-spatial pattern in the Drosophila embryo, based on a single well characterized enhancer model. The fly genome was searched to identify sequence elements containing the same combination of transcription factors as those found in the model. Experimental evaluation of the search results demonstrates that our method can correctly predict regulatory elements and highlights the importance of functional testing as a means of identifying false-positive results. We also show that the search results enable the identification of additional relevant sequence motifs whose functions can be empirically validated. This approach, combined with gene expression and phylogenetic sequence data, allows for genome-wide identification of related regulatory elements, an important step toward understanding the genetic regulatory networks involved in development.
Figures


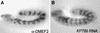


References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Arnone MI, Davidson EH. The hardwiring of development: Organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. - PubMed
-
- Blackman RK, Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986;188:499–515. - PubMed
-
- Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
