Alu-containing exons are alternatively spliced
- PMID: 12097342
- PMCID: PMC186627
- DOI: 10.1101/gr.229302
Alu-containing exons are alternatively spliced
Abstract
Alu repetitive elements are found in approximately 1.4 million copies in the human genome, comprising more than one-tenth of it. Numerous studies describe exonizations of Alu elements, that is, splicing-mediated insertions of parts of Alu sequences into mature mRNAs. To study the connection between the exonization of Alu elements and alternative splicing, we used a database of ESTs and cDNAs aligned to the human genome. We compiled two exon sets, one of 1176 alternatively spliced internal exons, and another of 4151 constitutively spliced internal exons. Sixty one alternatively spliced internal exons (5.2%) had a significant BLAST hit to an Alu sequence, but none of the constitutively spliced internal exons had such a hit. The vast majority (84%) of the Alu-containing exons that appeared within the coding region of mRNAs caused a frame-shift or a premature termination codon. Alu-containing exons were included in transcripts at lower frequencies than alternatively spliced exons that do not contain an Alu sequence. These results indicate that internal exons that contain an Alu sequence are predominantly, if not exclusively, alternatively spliced. Presumably, evolutionary events that cause a constitutive insertion of an Alu sequence into an mRNA are deleterious and selected against.
Figures

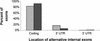
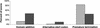

References
-
- Baban S, Freeman JD, Mager DL. Transcripts from a novel human KRAB zinc finger gene contain spliced Aluand endogenous retroviral segments. Genomics. 1996;33:463–472. - PubMed
-
- Batzer MA, Deininger PL, Hellmann-Blumberg U, Jurka J, Labuda D, Rubin CM, Schmid CW, Zietkiewicz E, Zuckerkandl E. Standardized nomenclature for Alurepeats. J Mol Evol. 1996;42:3–6. - PubMed
-
- Caras IW, Davitz MA, Rhee L, Weddell G, Martin DW, Jr, Nussenzweig V. Cloning of decay-accelerating factor suggests novel use of splicing to generate two proteins. Nature. 1987;325:545–549. - PubMed
-
- Claverie JM, Makalowski W. Alualert. Nature. 1994;371:752. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
