A computer-based method of selecting clones for a full-length cDNA project: simultaneous collection of negligibly redundant and variant cDNAs
- PMID: 12097351
- PMCID: PMC186622
- DOI: 10.1101/gr.75202
A computer-based method of selecting clones for a full-length cDNA project: simultaneous collection of negligibly redundant and variant cDNAs
Abstract
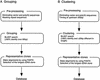
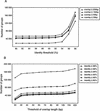
We describe a computer-based method that selects representative clones for full-length sequencing in a full-length cDNA project. Our method classifies end sequences using two kinds of criteria, grouping, and clustering. Grouping places together variant cDNAs, family genes, and cDNAs with sequencing errors. Clustering separates those cDNA clones into distinct clusters. The full-length sequences of the clones selected by grouping are determined preferentially, and then the sequences selected by clustering are determined. Grouping reduced the number of rice cDNA clones for full-length sequencing to 21% and mouse cDNA clones to 25%. Rice full-length sequences selected by grouping showed a 1.07-fold redundancy. Mouse full-length sequences showed a 1.04-fold redundancy, which can be reduced by approximately 30% from the selection using our previous method. To estimate the coverage of unique genes, we used FANTOM (Functional Annotation of RIKEN Mouse cDNA Clones) clusters (). Grouping covered almost all unique genes (93% of FANTOM clusters), and clustering covered all genes. Therefore, our method is useful for the selection of appropriate representative clones for full-length sequencing, thereby greatly reducing the cost, labor, and time necessary for this process.
Figures


References
-
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377 (6547 Suppl):3–174. - PubMed
-
- Boguski MS, Schuler GD. Establishing a human transcript map. Nat Genet. 1995;10:369–371. - PubMed
-
- Bouck J, Yu W, Gibbs R, Worley K. Comparison of gene indexing databases. Trends Genet. 1999;15:159–162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
