Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels
- PMID: 12097481
- PMCID: PMC3062512
- DOI: 10.1523/JNEUROSCI.22-13-05300.2002
Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels
Abstract
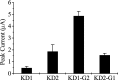
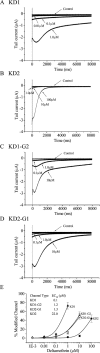
Alternative splicing is a major mechanism by which potassium and calcium channels increase functional diversity in animals. Extensive alternative splicing of the para sodium channel gene and developmental regulation of alternative splicing have been reported in Drosophila species. Alternative splicing has also been observed for several mammalian voltage-gated sodium channel genes. However, the functional significance of alternative splicing of sodium channels has not been demonstrated. In this study, we identified three mutually exclusive alternative exons encoding part of segments 3 and 4 of domain III in the German cockroach sodium channel gene, para(CSMA). The splice site is conserved in the mouse, fish, and human Na(v)1.6 sodium channel genes, suggesting an ancient origin. One of the alternative exons possesses a stop codon, which would generate a truncated protein with only the first two domains. The splicing variant containing the stop codon is detected only in the PNS, whereas the other two full-size variants were detected in both the PNS and CNS. When expressed in Xenopus oocytes, the two splicing variants produced robust sodium currents, but with different gating properties, whereas the splicing variant with the stop codon did not produce any detectable sodium current. Furthermore, these two functional splicing variants exhibited a striking difference in sensitivity to a pyrethroid insecticide, deltamethrin. Exon swapping partially reversed the channel sensitivity to deltamethrin. Our results therefore provide the first evidence that alternative splicing of a sodium channel gene produces pharmacologically distinct channels.
Figures

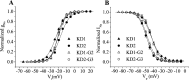

References
-
- Bell WL. The laboratory cockroach. Chapman and Hall; New York: 1981.
-
- Bourinet E, Soong TW, Sutton K, Slaymaker S, Mathews E, Monteil A, Zamponi GW, Nargeot J, Snutch TP. Splicing of alpha 1A subunit gene generates phenotypic variants of P- and Q. Nat Neurosci. 1999;2:407–415. - PubMed
-
- Burt PE, Goodchild RE. The site of action of pyrethrin I in the nervous system of the cockroach Periplaneta Americana. Ent Exp Appl. 1971;14:179–189.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
