Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway
- PMID: 12097557
- PMCID: PMC136367
- DOI: 10.1128/jvi.76.15.7453-7459.2002
Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway
Abstract
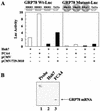
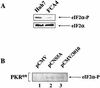
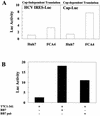
Hepatitis C virus (HCV) replicates from a ribonucleoprotein (RNP) complex that is associated with the endoplasmic reticulum (ER) membrane. The replication activities of the HCV subgenomic replicon are shown here to induce ER stress. In response to this stress, cells expressing HCV replicons induce the unfolded protein response (UPR), an ER-to-nucleus intracellular signaling pathway. The UPR is initiated by the proteolytic cleavage of a transmembrane protein, ATF6. The resulting cytoplasmic protein fragment of ATF6 functions as a transcription factor in the nucleus and activates selective genes required for an ER stress response. ATF6 activation leads to increased transcriptional levels of GRP78, an ER luminal chaperone protein. However, the overall level of GRP78 protein is decreased. While ER stress is also known to affect translational attenuation, cells expressing HCV replicons have lower levels of phosphorylation of the alpha subunit of eukaryotic initiation factor 2. Interestingly, cap-independent internal ribosome entry site-mediated translation directed by the 5' noncoding region of HCV and GRP78 is activated in cells expressing HCV replicons. These studies provide insight into the effects of HCV replication on intracellular events and the mechanisms underlying liver pathogenesis.
Figures
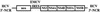


References
-
- Abraham, V., D. F. Stojkl, P. I. Duncan, N. Methot, T. Ishii, M. Dube, B. C. Vanderhyden, H. L. Atkins, D. A. Gray, M. W. McBurney, A. E. Koromilas, E. G. Brown, N. Sonenberg, and J. C. Bell. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 274:5953-5962. - PubMed
-
- Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. - PubMed
-
- Berlanga, J. J., J. Santoyo, and C. De Haro. 1999. Characterization of a mammalian homologue of the GCN2 eukaryotic initiation factor 2 alpha kinase. Eur. J. Biochem. 265:754-762. - PubMed
-
- Berry, M. J., G. S. Knutson, S. R. Lasky, S. M. Munemitsu, and C. E. Samuel. 1985. Mechanism of interferon from untreated and interferon-treated mouse fibroblasts. J. Biol. Chem. 270:11240-11247. - PubMed
-
- Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

