Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo
- PMID: 12097560
- PMCID: PMC136396
- DOI: 10.1128/jvi.76.15.7473-7484.2002
Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo
Abstract
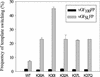
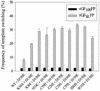
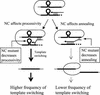
In vitro studies have indicated that retroviral nucleocapsid (NC) protein facilitates both DNA synthesis by reverse transcriptase (RT) and annealing of the nascent DNA with acceptor template. Increasing the rate of DNA synthesis is expected to reduce the frequency of RT template switching, whereas annealing the nascent DNA with acceptor template promotes template switching. We performed a mutational analysis of the murine leukemia virus (MLV) NC zinc finger domain to study its effect on RT template switching in vivo and to explore the role of NC during reverse transcription. The effects of NC mutations on RT template switching were determined by using a previously described in vivo direct-repeat deletion assay. A trans-complementation assay was also developed in which replication-defective NC mutants were rescued by coexpression of replication-defective RT mutants that provided wild-type NC in trans. We found that mutations in the MLV NC zinc finger domain increased the frequency of template switching approximately twofold. When a predicted stem-loop RNA secondary structure was introduced into the template RNA, the template-switching frequency increased 5-fold for wild-type NC and further increased up to an additional 6-fold for NC zinc finger domain mutants, resulting in an overall increase of as much as 30-fold. Thus, wild-type NC increased the efficiency with which RT was able to reverse transcribe through regions of RNA secondary structure that might serve as RT pause sites. These results provide the first in vivo evidence that NC enhances the rate of DNA synthesis by RT in regions of the template possessing stable RNA secondary structure.
Figures




Similar articles
-
HIV-1 nucleocapsid protein and the secondary structure of the binary complex formed between tRNA(Lys.3) and viral RNA template play different roles during initiation of (-) strand DNA reverse transcription.J Biol Chem. 2001 Dec 14;276(50):47725-32. doi: 10.1074/jbc.M105124200. Epub 2001 Oct 15. J Biol Chem. 2001. PMID: 11602578
-
Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue.J Virol. 1998 Nov;72(11):9034-44. doi: 10.1128/JVI.72.11.9034-9044.1998. J Virol. 1998. PMID: 9765448 Free PMC article.
-
The effect of template RNA structure on elongation by HIV-1 reverse transcriptase.Biochim Biophys Acta. 1999 Mar 19;1444(3):355-70. doi: 10.1016/s0167-4781(99)00011-1. Biochim Biophys Acta. 1999. PMID: 10095059
-
Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism.Prog Nucleic Acid Res Mol Biol. 2005;80:217-86. doi: 10.1016/S0079-6603(05)80006-6. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164976 Review. No abstract available.
-
Flexible nature and specific functions of the HIV-1 nucleocapsid protein.J Mol Biol. 2011 Jul 22;410(4):565-81. doi: 10.1016/j.jmb.2011.03.037. J Mol Biol. 2011. PMID: 21762801 Review.
Cited by
-
Rubella virus capsid protein modulates viral genome replication and virus infectivity.J Virol. 2004 Apr;78(8):4314-22. doi: 10.1128/jvi.78.8.4314-4322.2004. J Virol. 2004. PMID: 15047844 Free PMC article.
-
Reverse Transcription of Retroviruses and LTR Retrotransposons.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0027-2014. doi: 10.1128/microbiolspec.MDNA3-0027-2014. Microbiol Spectr. 2015. PMID: 26104704 Free PMC article. Review.
-
Charged assembly helix motif in murine leukemia virus capsid: an important region for virus assembly and particle size determination.J Virol. 2003 Jun;77(12):7058-66. doi: 10.1128/jvi.77.12.7058-7066.2003. J Virol. 2003. PMID: 12768025 Free PMC article.
-
The arginine clusters of the carboxy-terminal domain of the core protein of hepatitis B virus make pleiotropic contributions to genome replication.J Virol. 2011 Feb;85(3):1298-309. doi: 10.1128/JVI.01957-10. Epub 2010 Nov 17. J Virol. 2011. PMID: 21084467 Free PMC article.
-
Moloney murine leukemia virus genomic RNA packaged in the absence of a full complement of wild type nucleocapsid protein.Virology. 2012 Sep 1;430(2):100-9. doi: 10.1016/j.virol.2012.05.003. Epub 2012 May 25. Virology. 2012. PMID: 22633243 Free PMC article.
References
-
- Berkowitz, R. D., and S. P. Goff. 1994. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 202:233-246. - PubMed
-
- Buiser, R. G., R. A. Bambara, and P. J. Fay. 1993. Pausing by retroviral DNA polymerases promotes strand transfer from internal regions of RNA donor templates to homopolymeric acceptor templates. Biochim. Biophys. Acta 1216:20-30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

