Analysis of deletion mutants indicates that the 2A polypeptide of hepatitis A virus participates in virion morphogenesis
- PMID: 12097562
- PMCID: PMC136361
- DOI: 10.1128/jvi.76.15.7495-7505.2002
Analysis of deletion mutants indicates that the 2A polypeptide of hepatitis A virus participates in virion morphogenesis
Abstract
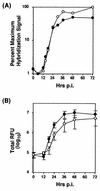
Unlike all other picornaviruses, the primary cleavage of the hepatitis A virus (HAV) polyprotein occurs at the 2A/2B junction and is carried out by the only proteinase encoded by the virus, 3C(pro). The resulting P1-2A capsid protein precursor is subsequently cleaved by 3C(pro) to generate VP0, VP3, and VP1-2A, which associate as pentamers. An unidentified cellular proteinase acting at the VP1/2A junction releases the mature capsid protein VP1 from VP1-2A later in the morphogenesis process. Although these aspects of polyprotein processing are well characterized, the function of 2A is unknown. To study its role in the viral life cycle, we assessed the infectivity of synthetic, genome-length RNAs containing 11 different in-frame deletions in the 2A region. Deletions in the N-terminal 40% of 2A abolished infectivity, whereas deletions in the C-terminal 60% resulted in viruses with a small-focus replication phenotype. C-terminal deletions in 2A had no effect on RNA replication kinetics under one-step growth conditions, nor did they have an effect on capsid protein synthesis and 3C(pro)-mediated processing. However, C-terminal deletions in 2A altered the VP1/2A cleavage, resulting in accumulation of uncleaved VP1-2A precursor in virions and possibly accounting for a delay in the appearance of infectious particles with these mutants, as well as a fourfold decrease in specific infectivity of the virus particles. When the capsid proteins were expressed from recombinant vaccinia viruses, the N-terminal part of 2A was required for efficient cleavage of the P1-2A precursor by 3C(pro) and assembly of structural precursors into pentamers. These data indicate that the N-terminal domain of 2A must be present as a C-terminal extension of P1 for folding of the capsid protein precursor to allow efficient 3C(pro)-mediated cleavages and to promote pentamer assembly, after which cleavage at the VP1/2A junction releases the mature VP1 protein, a process that appears to be necessary to produce highly infectious particles.
Figures







References
-
- Boege, U., and D. G. Scraba. 1989. Mengo virus maturation is accompanied by C-terminal modification of capsid protein VP1. Virology 168:409-412. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

