Comparison of predicted scaffold-compatible sequence variation in the triple-hairpin structure of human imunodeficiency virus type 1 gp41 with patient data
- PMID: 12097573
- PMCID: PMC136393
- DOI: 10.1128/jvi.76.15.7595-7606.2002
Comparison of predicted scaffold-compatible sequence variation in the triple-hairpin structure of human imunodeficiency virus type 1 gp41 with patient data
Abstract
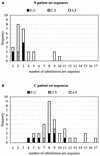
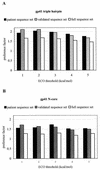
It has been proposed that the ectodomain of human immunodeficiency virus type 1 (HIV-1) gp41 (e-gp41), involved in HIV entry into the target cell, exists in at least two conformations, a pre-hairpin intermediate and a fusion-active hairpin structure. To obtain more information on the structure-sequence relationship in e-gp41, we performed in silico a full single-amino-acid substitution analysis, resulting in a Fold Compatible Database (FCD) for each conformation. The FCD contains for each residue position in a given protein a list of values assessing the energetic compatibility (ECO) of each of the 20 natural amino acids at that position. Our results suggest that FCD predictions are in good agreement with the sequence variation observed for well-validated e-gp41 sequences. The data show that at a minECO threshold value of 5 kcal/mol, about 90% of the observed patient sequence variation is encompassed by the FCD predictions. Some inconsistent FCD predictions at N-helix positions packing against residues of the C helix suggest that packing of both peptides may involve some flexibility and may be attributed to an altered orientation of the C-helical domain versus the N-helical region. The permissiveness of sequence variation in the C helices is in agreement with FCD predictions. Comparison of N-core and triple-hairpin FCDs suggests that the N helices may impose more constraints on sequence variation than the C helices. Although the observed sequences of e-gp41 contain many multiple mutations, our method, which is based on single-point mutations, can predict the natural sequence variability of e-gp41 very well.
Figures

References
-
- Beirnaert, E., P. Nyambi, B. Willems, L. Heyndrickx, B. Colebunders, W. Janssens, and G. van der Groen. 2000. Identification and characterization of sera from HIV-infected individuals with broad cross-neutralizing activity against group M (env clade A-H) and group O primary HIV-1 isolates. J. Med. Virol. 61:14-24. - PubMed
-
- Bernstein, F. C., T. F. Koetzle, G. J. B. Williams, E. F. Meywe, Jr., M. D. Brice, J. R. Rodgers, O. Kennard, T. Shimanoushi, and M. Tasumi. 1977. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 112:535-542. - PubMed
-
- Buzko, O. V., and K. M. Shokat. 1999. Blocking HIV entry. Nat. Struct. Biol. 6:906-908. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

