Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication
- PMID: 12097603
- PMCID: PMC136389
- DOI: 10.1128/jvi.76.15.7890-7896.2002
Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication
Abstract
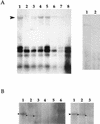
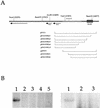
We used a transient-transfection replication assay to identify two functional copies of the human herpesvirus 8 (HHV8) lytic origin of DNA replication (oriLyt). BCLB-1 cells were transfected with HHV8 subgenomic fragments containing the putative lytic origin along with a plasmid expressing viral transactivator open reading frame (ORF) 50. The HHV8 left-end oriLyt (oriLyt-L) lies between ORFs K4.2 and K5 and is composed of a region encoding various transcription factor binding sites and an A+T-rich region and a G+C repeat region. The right-end oriLyt (oriLyt-R) maps between ORF 69 and vFLIP, a region similar to the RRV oriLyt, and is an inverted duplication of oriLyt-L.
Figures


References
-
- Birley, H. D., and T. F. Schultz. 1997. Kaposi's sarcoma and the new herpesvirus. J. Med. Microbiol. 46:433-435. - PubMed
-
- Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. - PubMed
-
- Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. - PubMed
-
- Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

