A complex prediction: three-dimensional model of the yeast exosome
- PMID: 12101094
- PMCID: PMC1084189
- DOI: 10.1093/embo-reports/kvf135
A complex prediction: three-dimensional model of the yeast exosome
Abstract
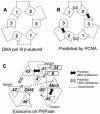
We present a model of the yeast exosome based on the bacterial degradosome component polynucleotide phosphorylase (PNPase). Electron microscopy shows the exosome to resemble PNPase but with key differences likely related to the position of RNA binding domains, and to the location of domains unique to the exosome. We use various techniques to reduce the many possible models of exosome subunits based on PNPase to just one. The model suggests numerous experiments to probe exosome function, particularly with respect to subunits making direct atomic contacts and conserved, possibly functional residues within the predicted central pore of the complex.
Figures


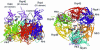
References
-
- Aloy P., Querol, E., Aviles, F.X. and Sternberg, M.J.E. (2001) Automated structure-based prediction of functional sites in proteins. J. Mol. Biol., 311, 395–408. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

