Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression
- PMID: 12101123
- PMCID: PMC186374
- DOI: 10.1101/gad.972202
Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression
Abstract
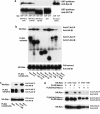
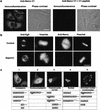
The protein kinase NIMA is an indispensable pleiotropic regulator of mitotic progression in Aspergillus. Although several mammalian NIMA-like kinases (Neks) are known, none appears to have the broad importance for mitotic regulation attributed to NIMA. Nercc1 is a new NIMA-like kinase that regulates chromosome alignment and segregation in mitosis. Its NIMA-like catalytic domain is followed by a noncatalytic tail containing seven repeats homologous to those of the Ran GEF, RCC1, a Ser/Thr/Pro-rich segment, and a coiled-coil domain. Nercc1 binds to another NIMA-like kinase, Nek6, and also binds specifically to the Ran GTPase through both its catalytic and its RCC1-like domains, preferring RanGDP in vivo. Nercc1 exists as a homooligomer and can autoactivate in vitro by autophosphorylation. Nercc1 is a cytoplasmic protein that is activated during mitosis and is avidly phosphorylated by active p34(Cdc2). Microinjection of anti-Nercc1 antibodies in prophase results in spindle abnormalities and/or chromosomal misalignment. In Ptk2 cells the outcome is prometaphase arrest or aberrant chromosome segregation and aneuploidy, whereas in CFPAC-1 cells prolonged arrest in prometaphase is the usual response. Nercc1 and its partner Nek6 represent a new signaling pathway that regulates mitotic progression.
Figures




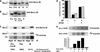

Similar articles
-
A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases.J Biol Chem. 2003 Sep 12;278(37):34897-909. doi: 10.1074/jbc.M303663200. Epub 2003 Jul 2. J Biol Chem. 2003. PMID: 12840024
-
The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation.J Cell Sci. 2008 Dec 1;121(Pt 23):3912-21. doi: 10.1242/jcs.035360. Epub 2008 Nov 11. J Cell Sci. 2008. PMID: 19001501 Free PMC article.
-
Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly.Mol Biol Cell. 2005 Oct;16(10):4827-40. doi: 10.1091/mbc.e05-04-0315. Epub 2005 Aug 3. Mol Biol Cell. 2005. PMID: 16079175 Free PMC article.
-
The NIMA kinase: a mitotic regulator in Aspergillus nidulans and vertebrate cells.Prog Cell Cycle Res. 1995;1:187-205. doi: 10.1007/978-1-4615-1809-9_15. Prog Cell Cycle Res. 1995. PMID: 9552363 Review.
-
Never say never. The NIMA-related protein kinases in mitotic control.Trends Cell Biol. 2003 May;13(5):221-8. doi: 10.1016/s0962-8924(03)00056-4. Trends Cell Biol. 2003. PMID: 12742165 Review.
Cited by
-
NIMA-related kinase 9 regulates the phosphorylation of the essential myosin light chain in the heart.Nat Commun. 2022 Oct 20;13(1):6209. doi: 10.1038/s41467-022-33658-2. Nat Commun. 2022. PMID: 36266340 Free PMC article.
-
Protein kinases of the Hippo pathway: regulation and substrates.Semin Cell Dev Biol. 2012 Sep;23(7):770-84. doi: 10.1016/j.semcdb.2012.07.002. Epub 2012 Aug 9. Semin Cell Dev Biol. 2012. PMID: 22898666 Free PMC article. Review.
-
Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of Pseudomitosis independent of US28 function.J Virol. 2004 Nov;78(21):11988-2011. doi: 10.1128/JVI.78.21.11988-12011.2004. J Virol. 2004. PMID: 15479839 Free PMC article.
-
Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity.Mol Biol Cell. 2003 Oct;14(10):4260-71. doi: 10.1091/mbc.e02-11-0773. Epub 2003 Jul 11. Mol Biol Cell. 2003. PMID: 14517334 Free PMC article.
-
Transcriptome analysis of a rotenone model of parkinsonism reveals complex I-tied and -untied toxicity mechanisms common to neurodegenerative diseases.PLoS One. 2012;7(9):e44700. doi: 10.1371/journal.pone.0044700. Epub 2012 Sep 7. PLoS One. 2012. PMID: 22970289 Free PMC article.
References
-
- Azuma Y, Renault L, Garcia-Ranea JA, Valencia A, Nishimoto T, Wittinghofer A. Model of the ran–RCC1 interaction using biochemical and docking experiments. J Mol Biol. 1999;289:1119–1130. - PubMed
-
- Belham C, Comb MJ, Avruch J. Identification of the NIMA family kinases NEK6/7 as regulators of the p70 ribosomal S6 kinase. Curr Biol. 2001;11:1155–1167. - PubMed
-
- Bischoff JR, Plowman GD. The Aurora/Ipl1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. - PubMed
-
- Borodovsky M, McIninch J. GeneMark: Parallel gene recognition for both DNA strands. Comput Chem. 1993;17:123–133.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
