Spo13 protects meiotic cohesin at centromeres in meiosis I
- PMID: 12101124
- PMCID: PMC186364
- DOI: 10.1101/gad.975802
Spo13 protects meiotic cohesin at centromeres in meiosis I
Abstract
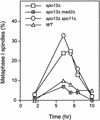
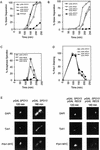
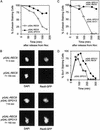
In the absence of Spo13, budding yeast cells complete a single meiotic division during which sister chromatids often separate. We investigated the function of Spo13 by following chromosomes tagged with green fluorescent protein. The occurrence of a single division in spo13Delta homozygous diploids depends on the spindle checkpoint. Eliminating the checkpoint accelerates meiosis I in spo13Delta cells and allows them to undergo two divisions in which sister chromatids often separate in meiosis I and segregate randomly in meiosis II. Overexpression of Spo13 and the meiosis-specific cohesin Rec8 in mitotic cells prevents separation of sister chromatids despite destruction of Pds1 and activation of Esp1. This phenotype depends on the combined overexpression of both proteins and mimics one aspect of meiosis I chromosome behavior. Overexpressing the mitotic cohesin, Scc1/Mcd1, does not substitute for Rec8, suggesting that the combined actions of Spo13 and Rec8 are important for preventing sister centromere separation in meiosis I.
Figures




Similar articles
-
An interplay between Shugoshin and Spo13 for centromeric cohesin protection and sister kinetochore mono-orientation during meiosis I in Saccharomyces cerevisiae.Curr Genet. 2018 Oct;64(5):1141-1152. doi: 10.1007/s00294-018-0832-x. Epub 2018 Apr 11. Curr Genet. 2018. PMID: 29644457
-
Spo13 regulates cohesin cleavage.Genes Dev. 2002 Jul 1;16(13):1672-81. doi: 10.1101/gad.989302. Genes Dev. 2002. PMID: 12101125 Free PMC article.
-
Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis.Curr Biol. 2004 Dec 29;14(24):2183-96. doi: 10.1016/j.cub.2004.12.020. Curr Biol. 2004. PMID: 15620645
-
Many functions of the meiotic cohesin.Chromosome Res. 2010 Dec;18(8):909-24. doi: 10.1007/s10577-010-9169-0. Epub 2010 Nov 18. Chromosome Res. 2010. PMID: 21086039 Review.
-
Shugoshin protects cohesin complexes at centromeres.Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):515-21, discussion 521. doi: 10.1098/rstb.2004.1607. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15897177 Free PMC article. Review.
Cited by
-
Meikin is a conserved regulator of meiosis-I-specific kinetochore function.Nature. 2015 Jan 22;517(7535):466-71. doi: 10.1038/nature14097. Epub 2014 Dec 24. Nature. 2015. PMID: 25533956
-
The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I.Genes Dev. 2005 Dec 15;19(24):3017-30. doi: 10.1101/gad.1373005. Genes Dev. 2005. PMID: 16357219 Free PMC article.
-
Rewiring of the phosphoproteome executes two meiotic divisions in budding yeast.EMBO J. 2024 Apr;43(7):1351-1383. doi: 10.1038/s44318-024-00059-8. Epub 2024 Feb 27. EMBO J. 2024. PMID: 38413836 Free PMC article.
-
The Spo13/Meikin pathway confines the onset of gamete differentiation to meiosis II in yeast.EMBO J. 2022 Feb 15;41(4):e109446. doi: 10.15252/embj.2021109446. Epub 2022 Jan 13. EMBO J. 2022. PMID: 35023198 Free PMC article.
-
An interplay between Shugoshin and Spo13 for centromeric cohesin protection and sister kinetochore mono-orientation during meiosis I in Saccharomyces cerevisiae.Curr Genet. 2018 Oct;64(5):1141-1152. doi: 10.1007/s00294-018-0832-x. Epub 2018 Apr 11. Curr Genet. 2018. PMID: 29644457
References
-
- Bernard P, Maure JF, Javerzat JP. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol. 2001;3:522–526. - PubMed
-
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 1999;19:463–472. - PubMed
-
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. - PubMed
-
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
