PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells
- PMID: 12101225
- PMCID: PMC133940
- DOI: 10.1128/MCB.22.15.5281-5295.2002
PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells
Abstract
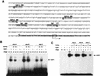
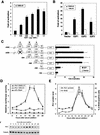
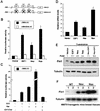
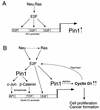
Oncogenes Neu/HER2/ErbB2 and Ras can induce mammary tumorigenesis via upregulation of cyclin D1. One major regulatory mechanism in these oncogenic signaling pathways is phosphorylation of serines or threonines preceding proline (pSer/Thr-Pro). Interestingly, the pSer/Thr-Pro motifs in proteins exist in two completely distinct cis and trans conformations, whose conversion is catalyzed specifically by the essential prolyl isomerase Pin1. By isomerizing pSer/Thr-Pro bonds, Pin1 can regulate the conformation and function of certain phosphorylated proteins. We have previously shown that Pin1 is overexpressed in breast tumors and positively regulates cyclin D1 by transcriptional activation and posttranslational stabilization. Moreover, in Pin1 knockout mice, mammary epithelial cells fail to undergo massive proliferation during pregnancy, as is the case in cyclin D1 null mice. These results indicate that Pin1 is upregulated in breast cancer and may be involved in mammary tumors. However, the mechanism of Pin1 overexpression in cancer and its significance in cell transformation remain largely unknown. Here we demonstrate that PIN1 expression is mediated by the transcription factor E2F and enhanced by c-Neu and Ha-Ras via E2F. Furthermore, overexpression of Pin1 not only confers transforming properties on mammary epithelial cells but also enhances the transformed phenotypes of Neu/Ras-transformed mammary epithelial cells. In contrast, inhibition of Pin1 suppresses Neu- and Ras-induced transformed phenotypes, which can be fully rescued by overexpression of a constitutively active cyclin D1 mutant that is refractory to the Pin1 inhibition. Thus, Pin1 is an E2F target gene that is essential for the Neu/Ras-induced transformation of mammary epithelial cells through activation of cyclin D1.
Figures
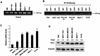
Similar articles
-
Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1.EMBO J. 2001 Jul 2;20(13):3459-72. doi: 10.1093/emboj/20.13.3459. EMBO J. 2001. PMID: 11432833 Free PMC article.
-
Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis.EMBO J. 2004 Aug 18;23(16):3397-407. doi: 10.1038/sj.emboj.7600323. Epub 2004 Jul 15. EMBO J. 2004. PMID: 15257284 Free PMC article.
-
Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes.Proc Natl Acad Sci U S A. 2002 Feb 5;99(3):1335-40. doi: 10.1073/pnas.032404099. Epub 2002 Jan 22. Proc Natl Acad Sci U S A. 2002. PMID: 11805292 Free PMC article.
-
The prolyl isomerase Pin1 in breast development and cancer.Breast Cancer Res. 2003;5(2):76-82. doi: 10.1186/bcr572. Epub 2003 Jan 28. Breast Cancer Res. 2003. PMID: 12631385 Free PMC article. Review.
-
Targeting carcinogenesis: a role for the prolyl isomerase Pin1?Mol Carcinog. 2006 Jun;45(6):397-402. doi: 10.1002/mc.20216. Mol Carcinog. 2006. PMID: 16652378 Review.
Cited by
-
SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function.Cancer Res. 2013 Jul 1;73(13):3951-62. doi: 10.1158/0008-5472.CAN-12-4360. Epub 2013 Apr 30. Cancer Res. 2013. PMID: 23633483 Free PMC article.
-
Stabilization of Pin1 by USP34 promotes Ubc9 isomerization and protein sumoylation in glioma stem cells.Nat Commun. 2024 Jan 2;15(1):40. doi: 10.1038/s41467-023-44349-x. Nat Commun. 2024. PMID: 38167292 Free PMC article.
-
Degradation of the tumor suppressor PML by Pin1 contributes to the cancer phenotype of breast cancer MDA-MB-231 cells.Mol Cell Biol. 2008 Feb;28(3):997-1006. doi: 10.1128/MCB.01848-07. Epub 2007 Nov 26. Mol Cell Biol. 2008. PMID: 18039859 Free PMC article.
-
Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function.J Virol. 2006 May;80(9):4276-85. doi: 10.1128/JVI.80.9.4276-4285.2006. J Virol. 2006. PMID: 16611886 Free PMC article.
-
PIN1 promoter polymorphism (-842 G>C) contributes to a decreased risk of cancer: Evidence from meta-analysis.Oncol Lett. 2014 Sep;8(3):1360-1366. doi: 10.3892/ol.2014.2280. Epub 2014 Jun 24. Oncol Lett. 2014. PMID: 25120724 Free PMC article.
References
-
- Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. - PubMed
-
- Arber, N., Y. Doki, E. K. Han, A. Sgambato, P. Zhou, N. H. Kim, T. Delohery, M. G. Klein, P. R. Holt, and I. B. Weinstein. 1997. Antisense to cyclin D1 inhibits the growth and tumorigenicity of human colon cancer cells. Cancer Res. 57:1569-1574. - PubMed
-
- Bartkova, J., J. Lukas, H. Muller, D. Lutzhoft, M. Strauss, and J. Bartek. 1994. Cyclin D1 protein expression and function in human breast cancer. Int. J. Cancer 57:353-361. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
