Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound
- PMID: 12101242
- PMCID: PMC133949
- DOI: 10.1128/MCB.22.15.5492-5505.2002
Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound
Abstract
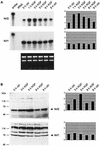
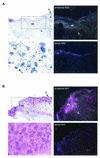
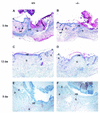
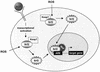
Keratinocyte growth factor (KGF) is a potent mitogen for epithelial cells, and it promotes survival of these cells under stress conditions. In a search for KGF-regulated genes in keratinocytes, we identified the gene encoding the transcription factor NF-E2-related factor 2 (Nrf2). Nrf2 is a key player in the cellular stress response. This might be of particular importance during wound healing, where large amounts of reactive oxygen species are produced as a defense against invading bacteria. Therefore, we studied the wound repair process in Nrf2 knockout mice. Interestingly, the expression of various key players involved in wound healing was significantly reduced in early wounds of the Nrf2 knockout animals, and the late phase of repair was characterized by prolonged inflammation. However, these differences in gene expression were not reflected by obvious histological abnormalities. The normal healing rate appears to be at least partially due to an up-regulation of the related transcription factor Nrf3, which was also identified as a target of KGF and which was coexpressed with Nrf2 in the healing skin wound. Taken together, our results reveal novel roles of the KGF-regulated transcription factors Nrf2 and possibly Nrf3 in the control of gene expression and inflammation during cutaneous wound repair.
Figures







Similar articles
-
Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages.J Biol Chem. 2000 May 26;275(21):16023-9. doi: 10.1074/jbc.275.21.16023. J Biol Chem. 2000. PMID: 10821856
-
Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation.J Biol Chem. 2001 Jun 15;276(24):20858-65. doi: 10.1074/jbc.M101198200. Epub 2001 Mar 26. J Biol Chem. 2001. PMID: 11274184
-
Caveolin-1 and -2 expression is differentially regulated in cultured keratinocytes and within the regenerating epidermis of cutaneous wounds.Exp Cell Res. 2000 Jul 10;258(1):23-32. doi: 10.1006/excr.2000.4904. Exp Cell Res. 2000. PMID: 10912784
-
Nrf2 Activation in Keratinocytes: A Central Role in Diabetes-Associated Wound Healing.Exp Dermatol. 2024 Oct;33(10):e15189. doi: 10.1111/exd.15189. Exp Dermatol. 2024. PMID: 39373525 Review.
-
[Molecular biology contributions to wound healing and practical applications].Langenbecks Arch Chir Suppl Kongressbd. 1998;115:678-82. Langenbecks Arch Chir Suppl Kongressbd. 1998. PMID: 9931700 Review. German.
Cited by
-
The KEAP1/NRF2 Signaling Pathway in Keratinization.Antioxidants (Basel). 2020 Aug 14;9(8):751. doi: 10.3390/antiox9080751. Antioxidants (Basel). 2020. PMID: 32823937 Free PMC article. Review.
-
Soshiho-Tang, a Traditional Herbal Medicine, Alleviates Atopic Dermatitis Symptoms via Regulation of Inflammatory Mediators.Front Pharmacol. 2019 Jul 3;10:742. doi: 10.3389/fphar.2019.00742. eCollection 2019. Front Pharmacol. 2019. PMID: 31338033 Free PMC article.
-
Peroxiredoxin 6 is required for blood vessel integrity in wounded skin.J Cell Biol. 2007 Nov 19;179(4):747-60. doi: 10.1083/jcb.200706090. J Cell Biol. 2007. PMID: 18025307 Free PMC article.
-
Protective Effects of Cannabidiol on Chemotherapy-Induced Oral Mucositis via the Nrf2/Keap1/ARE Signaling Pathways.Oxid Med Cell Longev. 2022 May 25;2022:4619760. doi: 10.1155/2022/4619760. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35669853 Free PMC article.
-
The Role of Mitophagy in Innate Immunity.Front Immunol. 2018 Jun 5;9:1283. doi: 10.3389/fimmu.2018.01283. eCollection 2018. Front Immunol. 2018. PMID: 29951054 Free PMC article. Review.
References
-
- Beer, H. D., C. Florence, J. Dammeier, L. McGuire, S. Werner, and D. R. Duan. 1997. Mouse fibroblast growth factor 10: cDNA cloning, protein characterization, and regulation of mRNA expression. Oncogene 15:2211-2218. - PubMed
-
- Bloch, W., K. Huggel, R. Sasaki, R. Grose, P. Bugnon, K. Addicks, R. Timpl, and S. Werner. 2000. The angiogenesis inhibitor endostatin impairs blood vessel maturation during wound healing. FASEB J. 14:2373-2376. - PubMed
-
- Caldelari, R., M. M. Suter, D. Baumann, A. De Bruin, and E. Müller. 2000. Long-term culture of murine epidermal keratinocytes. J. Investig. Dermatol. 114:1064-1065. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
