Major histocompatibility complex class II transcriptional platform: assembly of nuclear factor Y and regulatory factor X (RFX) on DNA requires RFX5 dimers
- PMID: 12101253
- PMCID: PMC133954
- DOI: 10.1128/MCB.22.15.5616-5625.2002
Major histocompatibility complex class II transcriptional platform: assembly of nuclear factor Y and regulatory factor X (RFX) on DNA requires RFX5 dimers
Abstract
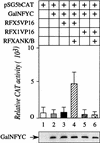
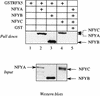
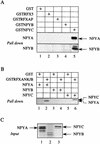
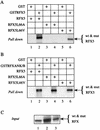
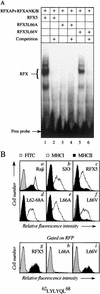
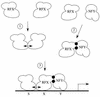
Major histocompatibility complex class II (MHC-II) genes are regulated in a B-cell-specific and gamma interferon-inducible manner. Conserved upstream sequences (CUS) in their compact promoters bind nuclear factor Y (NFY) and regulatory factor X (RFX) complexes. These DNA-bound proteins form a platform that attracts the class II transactivator, which initiates and elongates MHC-II transcription. In this report, we analyzed the complex assembly of these DNA-bound proteins. First, we found that NFY can interact with RFX in cells. In particular, NFYA and NFYC bound RFXANK/B in vitro. Next, RFX5 formed dimers in vivo and in vitro. Within a leucine-rich stretch N-terminal to the DNA-binding domain in RFX5, the leucine at position 66 was found to be critical for this self-association. Mutant RFX5 proteins that could not form dimers also did not support the formation of higher-order DNA-protein complexes on CUS in vitro or MHC-II transcription in vivo. We conclude that the MHC-II transcriptional platform begins to assemble off CUS and then binds DNA via multiple, spatially constrained interactions. These findings offer one explanation of why in the Bare Lymphocyte Syndrome, which is a congenital severe combined immunodeficiency, MHC-II promoters are bare when any subunit of RFX is mutated or missing.
Figures



Similar articles
-
A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y.Mol Cell Biol. 2000 May;20(10):3364-76. doi: 10.1128/MCB.20.10.3364-3376.2000. Mol Cell Biol. 2000. PMID: 10779326 Free PMC article.
-
Assembly of major histocompatibility complex (MHC) class II transcription factors: association and promoter recognition of RFX proteins.Biochemistry. 2004 Oct 12;43(40):12750-60. doi: 10.1021/bi030262o. Biochemistry. 2004. PMID: 15461447
-
MHC class II enhanceosome: how is the class II transactivator recruited to DNA-bound activators?Int Immunol. 2003 Apr;15(4):467-75. doi: 10.1093/intimm/dxg048. Int Immunol. 2003. PMID: 12663676
-
The bare lymphocyte syndrome and the regulation of MHC expression.Annu Rev Immunol. 2001;19:331-73. doi: 10.1146/annurev.immunol.19.1.331. Annu Rev Immunol. 2001. PMID: 11244040 Review.
-
Molecular genetics of the Bare lymphocyte syndrome.Rev Immunogenet. 2000;2(2):267-82. Rev Immunogenet. 2000. PMID: 11258423 Review.
Cited by
-
What is the role of alternate splicing in antigen presentation by major histocompatibility complex class I molecules?Immunol Res. 2010 Mar;46(1-3):32-44. doi: 10.1007/s12026-009-8123-8. Immunol Res. 2010. PMID: 19830395 Free PMC article. Review.
-
Interferon-gamma induces major histocompatibility class II transactivator (CIITA), which mediates collagen repression and major histocompatibility class II activation by human aortic smooth muscle cells.Circ Res. 2006 Mar 3;98(4):472-9. doi: 10.1161/01.RES.0000204725.46332.97. Epub 2006 Jan 26. Circ Res. 2006. PMID: 16439692 Free PMC article.
-
CpG methylation at the USF-binding site mediates cell-specific transcription of human ascorbate transporter SVCT2 exon 1a.Biochem J. 2011 Nov 15;440(1):73-84. doi: 10.1042/BJ20110392. Biochem J. 2011. PMID: 21770893 Free PMC article.
-
NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression.J Immunol. 2012 May 15;188(10):4951-8. doi: 10.4049/jimmunol.1103160. Epub 2012 Apr 6. J Immunol. 2012. PMID: 22490869 Free PMC article.
-
Regulatory factor X transcription factors control Musashi1 transcription in mouse neural stem/progenitor cells.Stem Cells Dev. 2014 Sep 15;23(18):2250-61. doi: 10.1089/scd.2014.0219. Stem Cells Dev. 2014. PMID: 25058468 Free PMC article.
References
-
- Alberti, S., S. Oehler, B. von Wilcken-Bergmann, H. Kramer, and B. Muller-Hill. 1991. Dimer-to-tetramer assembly of Lac repressor involves a leucine heptad repeat. New Biol. 3:57-62. - PubMed
-
- Boss, J. M. 1999. A common set of factors control the expression of the MHC class II, invariant chain, and HLA-DM genes. Microbes Infect. 1:847-853. - PubMed
-
- Bottazzo, G. F., R. Pujol-Borrell, T. Hanafusa, and M. Feldmann. 1983. Role of aberrant HLA-DR expression and antigen presentation in induction of endocrine autoimmunity. Lancet ii:1115-1119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
