Interaction of the Yersinia pestis type III regulatory proteins LcrG and LcrV occurs at a hydrophobic interface
- PMID: 12102728
- PMCID: PMC117220
- DOI: 10.1186/1471-2180-2-16
Interaction of the Yersinia pestis type III regulatory proteins LcrG and LcrV occurs at a hydrophobic interface
Erratum in
- BMC Microbiol. 2002 Sep 5;2(1):25.
Abstract
Background: Secretion of anti-host proteins by Yersinia pestis via a type III mechanism is not constitutive. The process is tightly regulated and secretion occurs only after an appropriate signal is received. The interaction of LcrG and LcrV has been demonstrated to play a pivotal role in secretion control. Previous work has shown that when LcrG is incapable of interacting with LcrV, secretion of anti-host proteins is prevented. Therefore, an understanding of how LcrG interacts with LcrV is required to evaluate how this interaction regulates the type III secretion system of Y. pestis. Additionally, information about structure-function relationships within LcrG is necessary to fully understand the role of this key regulatory protein.
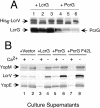
Results: In this study we demonstrate that the N-terminus of LcrG is required for interaction with LcrV. The interaction likely occurs within a predicted amphipathic coiled-coil domain within LcrG. Our results demonstrate that the hydrophobic face of the putative helix is required for LcrV interaction. Additionally, we demonstrate that the LcrG homolog, PcrG, is incapable of blocking type III secretion in Y. pestis. A genetic selection was utilized to obtain a PcrG variant capable of blocking secretion. This PcrG variant allowed us to locate a region of LcrG involved in secretion blocking.
Conclusion: Our results demonstrate that LcrG interacts with LcrV via hydrophobic interactions located in the N-terminus of LcrG within a predicted coiled-coil motif. We also obtained preliminary evidence that the secretion blocking activity of LcrG is located between amino acids 39 and 53.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

