The Tnfrh1 (Tnfrsf23) gene is weakly imprinted in several organs and expressed at the trophoblast-decidua interface
- PMID: 12102730
- PMCID: PMC117226
- DOI: 10.1186/1471-2156-3-11
The Tnfrh1 (Tnfrsf23) gene is weakly imprinted in several organs and expressed at the trophoblast-decidua interface
Abstract
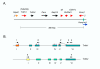
Background: The Tnfrh1 gene (gene symbol Tnfrsf23) is located near one end of a megabase-scale imprinted region on mouse distal chromosome 7, about 350 kb distant from the nearest known imprinting control element. Within 20 kb of Tnfrh1 is a related gene called Tnfrh2 (Tnfrsf22) These duplicated genes encode putative decoy receptors in the tumor necrosis factor (TNF) receptor family. Although other genes in this chromosomal region show conserved synteny with genes on human Chr11p15.5, there are no obvious human orthologues of Tnfrh1 or Tnfrh2.
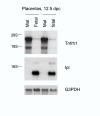
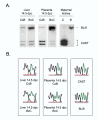
Results: We analyzed Tnfrh1 for evidence of parental imprinting, and characterized its tissue-specific expression. Tnfrh1 mRNA is detectable in multiple adult and fetal tissues, with highest expression in placenta, where in situ hybridization reveals a distinctive population of Tnfrh1-positive cells in maternal decidua, directly beneath the trophoblast giant cells. In offspring of interspecific mouse crosses, Tnfrh1 shows a consistent parent-of-origin-dependent allelic expression bias, with relative repression, but not silencing, of the paternal allele in several organs including fetal liver and adult spleen.
Conclusions: Genes preferentially expressed in the placenta are predicted to evolve rapidly, and Tnfrh1 appears to be an example of this phenomenon. In view of its strong expression in cells at the fetal-maternal boundary, Tnfrh1 warrants further study as a gene that might modulate immune or trophic interactions between the invasive placental trophoblast and the maternal decidua. The preferential expression of Tnfrh1 from the maternal allele indicates weak functional imprinting of this locus in some tissues.
Figures
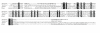




Similar articles
-
Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression.Hum Mol Genet. 2012 Feb 1;21(3):548-58. doi: 10.1093/hmg/ddr488. Epub 2011 Oct 24. Hum Mol Genet. 2012. PMID: 22025075
-
IMPT1, an imprinted gene similar to polyspecific transporter and multi-drug resistance genes.Hum Mol Genet. 1998 Apr;7(4):597-608. doi: 10.1093/hmg/7.4.597. Hum Mol Genet. 1998. PMID: 9499412
-
Tumor necrosis factor-alpha mRNA-positive cells in spontaneous resorption in rodents.Am J Reprod Immunol. 1998 Jan;39(1):50-7. doi: 10.1111/j.1600-0897.1998.tb00333.x. Am J Reprod Immunol. 1998. PMID: 9458934
-
Imprinting of the mouse Igf2r gene depends on an intronic CpG island.Mol Cell Endocrinol. 1998 May 25;140(1-2):9-14. doi: 10.1016/s0303-7207(98)00022-7. Mol Cell Endocrinol. 1998. PMID: 9722161 Review.
-
The placental imprintome and imprinted gene function in the trophoblast glycogen cell lineage.Reprod Biomed Online. 2012 Jul;25(1):44-57. doi: 10.1016/j.rbmo.2012.03.019. Epub 2012 Apr 4. Reprod Biomed Online. 2012. PMID: 22560119 Free PMC article. Review.
Cited by
-
A genomic reservoir for Tnfrsf genes is developmentally regulated and imprinted in the mouse.Epigenetics. 2012 Jun 1;7(6):626-34. doi: 10.4161/epi.20243. Epub 2012 Jun 1. Epigenetics. 2012. PMID: 22595876 Free PMC article.
-
Chronic airway-induced allergy in mice modifies gene expression in the brain toward insulin resistance and inflammatory responses.J Neuroinflammation. 2013 Aug 1;10:99. doi: 10.1186/1742-2094-10-99. J Neuroinflammation. 2013. PMID: 23915208 Free PMC article.
-
Biallelic expression of HRAS and MUCDHL in human and mouse.Hum Genet. 2003 Apr;112(4):334-42. doi: 10.1007/s00439-003-0907-7. Epub 2003 Feb 14. Hum Genet. 2003. PMID: 12589428
-
Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6.Genome Res. 2003 Jul;13(7):1696-705. doi: 10.1101/gr.906803. Genome Res. 2003. PMID: 12840045 Free PMC article.
-
Placental effects on the maternal brain revealed by disrupted placental gene expression in mouse hybrids.Proc Biol Sci. 2020 Jan 15;287(1918):20192563. doi: 10.1098/rspb.2019.2563. Epub 2020 Jan 15. Proc Biol Sci. 2020. PMID: 31937228 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

