Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors
- PMID: 12107135
- PMCID: PMC135211
- DOI: 10.1128/JB.184.15.4177-4186.2002
Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors
Erratum in
- J Bacteriol. 2003 Feb;185(4):1484
Abstract
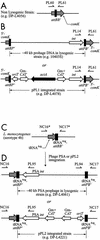
Two site-specific shuttle integration vectors were developed with two different chromosomal bacteriophage integration sites to facilitate strain construction in Listeria monocytogenes. The first vector, pPL1, utilizes the listeriophage U153 integrase and attachment site within the comK gene for chromosomal insertion. pPL1 contains a useful polylinker, can be directly conjugated from Escherichia coli into L. monocytogenes, forms stable, single-copy integrants at a frequency of approximately 10(-4) per donor cell, and can be used in the L. monocytogenes 1/2 and 4b serogroups. Methods for curing endogenous prophages from the comK attachment site in 10403S-derived strains were developed. pPL1 was used to introduce the hly and actA genes at comK-attBB' in deletion strains derived from 10403S and SLCC-5764. These strains were tested for second-site complementation in hemolysin assays, plaquing assays, and cell extract motility assays. Unlike plasmid-complemented strains, integrated pPL1-complemented strains were fully virulent in the mouse 50% lethal dose assay. Additionally, the PSA phage attachment site on the L. monocytogenes chromosome was characterized, and pPL1 was modified to integrate at this site. The listeriophage PSA integrates in the 3' end of an arginine tRNA gene. There are 17 bp of DNA identity between the bacterial and phage attachment sites. The PSA prophage DNA sequence reconstitutes a complete tRNA(Arg) gene. The modified vector, pPL2, was integration proficient at the same frequency as pPL1 in common laboratory serotype 1/2 strains as well as serotype 4b strains.
Figures



References
-
- Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

