Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds
- PMID: 12107280
- PMCID: PMC124975
- DOI: 10.1073/pnas.142310499
Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds
Abstract
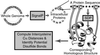
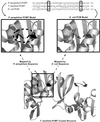
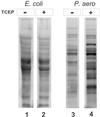
Disulfide bonds have only rarely been found in intracellular proteins. That pattern is consistent with the chemically reducing environment inside the cells of well-studied organisms. However, recent experiments and new calculations based on genomic data of archaea provide striking contradictions to this pattern. Our results indicate that the intracellular proteins of certain hyperthermophilic archaea, especially the crenarchaea Pyrobaculum aerophilum and Aeropyrum pernix, are rich in disulfide bonds. This finding implicates disulfide bonding in stabilizing many thermostable proteins and points to novel chemical environments inside these microbes. These unexpected results illustrate the wealth of biochemical insights available from the growing reservoir of genomic data.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

