Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii
- PMID: 12110581
- PMCID: PMC126117
- DOI: 10.1093/emboj/cdf372
Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii
Abstract
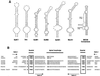
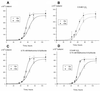
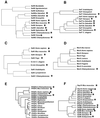
Known eukaryotic selenocysteine (Sec)-containing proteins are animal proteins, whereas selenoproteins have not been found in yeast and plants. Surprisingly, we detected selenoproteins in a member of the plant kingdom, Chlamydomonas reinhardtii, and directly identified two of them as phospholipid hydroperoxide glutathione peroxidase and selenoprotein W homologs. Moreover, a selenocysteyl-tRNA was isolated that recognized specifically the Sec codon UGA. Subsequent gene cloning and bioinformatics analyses identified eight additional selenoproteins, including methionine-S-sulfoxide reductase, a selenoprotein specific to Chlamydomonas: Chlamydomonas selenoprotein genes contained selenocysteine insertion sequence (SECIS) elements that were similar, but not identical, to those of animals. These SECIS elements could direct selenoprotein synthesis in mammalian cells, indicating a common origin of plant and animal Sec insertion systems. We found that selenium is required for optimal growth of Chlamydomonas: Finally, evolutionary analyses suggested that selenoproteins present in Chlamydomonas and animals evolved early, and were independently lost in land plants, yeast and some animals.
Figures







Similar articles
-
Selenium metabolism in zebrafish: multiplicity of selenoprotein genes and expression of a protein containing 17 selenocysteine residues.Genes Cells. 2000 Dec;5(12):1049-60. doi: 10.1046/j.1365-2443.2000.00392.x. Genes Cells. 2000. PMID: 11168591
-
High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes.J Mol Biol. 1999 Oct 8;292(5):1003-16. doi: 10.1006/jmbi.1999.3085. J Mol Biol. 1999. PMID: 10512699
-
Characterization of the UGA-recoding and SECIS-binding activities of SECIS-binding protein 2.RNA Biol. 2014;11(11):1402-13. doi: 10.1080/15476286.2014.996472. RNA Biol. 2014. PMID: 25692238 Free PMC article.
-
Selenoprotein synthesis in archaea.Biofactors. 2001;14(1-4):75-83. doi: 10.1002/biof.5520140111. Biofactors. 2001. PMID: 11568443 Review.
-
Selenium. Role of the essential metalloid in health.Met Ions Life Sci. 2013;13:499-534. doi: 10.1007/978-94-007-7500-8_16. Met Ions Life Sci. 2013. PMID: 24470102 Free PMC article. Review.
Cited by
-
Impact of oxidative stress on ascorbate biosynthesis in Chlamydomonas via regulation of the VTC2 gene encoding a GDP-L-galactose phosphorylase.J Biol Chem. 2012 Apr 20;287(17):14234-45. doi: 10.1074/jbc.M112.341982. Epub 2012 Mar 5. J Biol Chem. 2012. PMID: 22393048 Free PMC article.
-
Plant methionine sulfoxide reductase A and B multigenic families.Photosynth Res. 2006 Sep;89(2-3):247-62. doi: 10.1007/s11120-006-9097-1. Epub 2006 Sep 22. Photosynth Res. 2006. PMID: 17031545 Review.
-
Alternative first exon splicing regulates subcellular distribution of methionine sulfoxide reductases.BMC Mol Biol. 2006 Mar 16;7:11. doi: 10.1186/1471-2199-7-11. BMC Mol Biol. 2006. PMID: 16542431 Free PMC article.
-
A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells.Proc Natl Acad Sci U S A. 2007 May 8;104(19):7857-62. doi: 10.1073/pnas.0610683104. Epub 2007 Apr 30. Proc Natl Acad Sci U S A. 2007. PMID: 17470795 Free PMC article.
-
Bioaccumulation and toxicity of selenium compounds in the green alga Scenedesmus quadricauda.BMC Plant Biol. 2009 May 15;9:58. doi: 10.1186/1471-2229-9-58. BMC Plant Biol. 2009. PMID: 19445666 Free PMC article.
References
-
- Arner E.S., Sarioglu,H., Lottspeich,F., Holmgren,A. and Bock,A. (1999) High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J. Mol. Biol., 292, 1003–1016. - PubMed
-
- Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) Dynamics of the genetic code. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–673.
-
- Bock A. (2001) Selenium metabolism in bacteria. In Hatfield,D.L. (ed.), Selenium: Its Molecular Biology and Role in Human Health. Kluwer Academic, Norwell, MA, pp. 7–22.
-
- Bock A., Forchhammer,K., Heider,J. and Baron,C. (1991) Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem. Sci., 16, 463–467. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

