Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP
- PMID: 12110593
- PMCID: PMC126121
- DOI: 10.1093/emboj/cdf376
Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP
Abstract
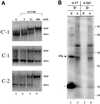
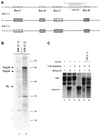
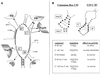
Methylation of the ribose 2'-hydroxyl, the most widespread modification of ribosomal and splicesomal RNAs, is guided by the box C/D class of small nucleolar RNAs (snoRNAs). Box C/D small nucleolar ribonucleoproteins (snoRNPs) contain four core proteins: fibrillarin, Nop56, Nop58 and 15.5 kDa. We constructed U25 snoRNAs containing a single photoactivatable 4-thiouridine at each U position within the conserved box C/D and C'/D' motifs. Proteins assembled on the snoRNA after injection into Xenopus oocyte nuclei were identified by cross-linking, and reconstituted particles characterized by functional rescue and mutational analyses. Our data argue that box C/D snoRNPs are asymmetric, with the C' box contacting Nop56 and fibrillarin, the C box interacting with Nop58, and the D and D' boxes contacting fibrillarin. No cross-link to 15.5 kDa was detected; its binding is disrupted by 4-thiouridine substitution in position 1 of the C box. Repositioning the guide sequence of U25 upstream of box D instead of D' revealed that both C/D motifs have the potential to function as guide centers, but, surprisingly, there was no alteration in protein cross-linking.
Figures


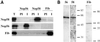
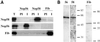



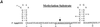
References
-
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. - PubMed
-
- Cavaillé J., Nicoloso,M. and Bachellerie,J.P. (1996) Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature, 383, 732–735. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

