Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition
- PMID: 12110594
- PMCID: PMC126118
- DOI: 10.1093/emboj/cdf373
Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition
Abstract
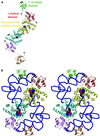
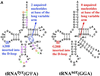
Bacterial tyrosyl-tRNA synthetases (TyrRS) possess a flexibly linked C-terminal domain of approximately 80 residues, which has hitherto been disordered in crystal structures of the enzyme. We have determined the structure of Thermus thermophilus TyrRS at 2.0 A resolution in a crystal form in which the C-terminal domain is ordered, and confirm that the fold is similar to part of the C-terminal domain of ribosomal protein S4. We have also determined the structure at 2.9 A resolution of the complex of T.thermophilus TyrRS with cognate tRNA(tyr)(G Psi A). In this structure, the C-terminal domain binds between the characteristic long variable arm of the tRNA and the anti-codon stem, thus recognizing the unique shape of the tRNA. The anticodon bases have a novel conformation with A-36 stacked on G-34, and both G-34 and Psi-35 are base-specifically recognized. The tRNA binds across the two subunits of the dimeric enzyme and, remarkably, the mode of recognition of the class I TyrRS for its cognate tRNA resembles that of a class II synthetase in being from the major groove side of the acceptor stem.
Figures
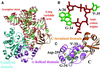




Similar articles
-
tRNA(Pro) anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase.Structure. 1998 Jan 15;6(1):101-8. doi: 10.1016/s0969-2126(98)00011-2. Structure. 1998. PMID: 9493271
-
Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion.Nat Struct Biol. 2003 Jun;10(6):425-32. doi: 10.1038/nsb934. Nat Struct Biol. 2003. PMID: 12754495
-
The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe.Structure. 1997 Jan 15;5(1):59-68. doi: 10.1016/s0969-2126(97)00166-4. Structure. 1997. PMID: 9016717
-
Evolution of the tRNA(Tyr)/TyrRS aminoacylation systems.Biochimie. 2005 Sep-Oct;87(9-10):873-83. doi: 10.1016/j.biochi.2005.03.008. Epub 2005 Apr 8. Biochimie. 2005. PMID: 16164994 Review.
-
Discrimination between transfer-RNAs by tyrosyl-tRNA synthetase.Biochimie. 1993;75(12):1099-108. doi: 10.1016/0300-9084(93)90009-h. Biochimie. 1993. PMID: 8199245 Review.
Cited by
-
The RNA degradosome promotes tRNA quality control through clearance of hypomodified tRNA.Proc Natl Acad Sci U S A. 2019 Jan 22;116(4):1394-1403. doi: 10.1073/pnas.1814130116. Epub 2019 Jan 8. Proc Natl Acad Sci U S A. 2019. PMID: 30622183 Free PMC article.
-
Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures.Int J Mol Sci. 2018 Dec 26;20(1):92. doi: 10.3390/ijms20010092. Int J Mol Sci. 2018. PMID: 30587834 Free PMC article.
-
The double-length tyrosyl-tRNA synthetase from the eukaryote Leishmania major forms an intrinsically asymmetric pseudo-dimer.J Mol Biol. 2011 Jun 3;409(2):159-76. doi: 10.1016/j.jmb.2011.03.026. Epub 2011 Mar 21. J Mol Biol. 2011. PMID: 21420975 Free PMC article.
-
Conformational landscapes for KMSKS loop in tyrosyl-tRNA synthetases.J Struct Funct Genomics. 2014 Jun;15(2):45-61. doi: 10.1007/s10969-014-9178-x. Epub 2014 Apr 11. J Struct Funct Genomics. 2014. PMID: 24723074
-
The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity.J Biol Chem. 2008 Aug 8;283(32):21997-2006. doi: 10.1074/jbc.M801838200. Epub 2008 Jun 17. J Biol Chem. 2008. PMID: 18559342 Free PMC article.
References
-
- Aravind L. and Koonin,E.V. (1999) Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol., 48, 291–302. - PubMed
-
- Asahara H., Himeno,H., Tamura,K., Hasegawa,T., Watanabe,K. and Shimizu,M. (1993) Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J. Mol. Biol., 231, 219–229. - PubMed
-
- Bedouelle H. (1990) Recognition of tRNAtyr by tyrosyl-tRNA synthetase. Biochimie, 72, 589–598. - PubMed
-
- Bedouelle H., Guez-Ivanier,V. and Nageotte,R. (1993) Discrimination between transfer-RNAs by tyrosyl-tRNA synthetase. Biochimie, 75, 1099–1108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

